Restriction endonuclease enzyme is also known as restriction enzyme or restrictase or molecular scissors which is a type of protein produced by bacteria that cleaves at or near specific restriction site along the molecule.
Werner Arber and Matthew Meselson originated the restriction enzyme from the Enterobacteria lambda phage. Certain strain of E. coli has an ability to inhibit the activity of lambda phage by the enzymatic cleavage. Their discovery and characterization of restriction enzymes led to the development of recombinant DNA technology.
How restriction enzyme works?
Restriction enzymes can be isolated from the bacterial or archeal cell to manipulate fragments of DNA in the laboratory for recombinant DNA technology and genetic engineering.
Bacterial cell cleaves foreign DNA with the help of restriction enzyme which defend and eliminates infecting organisms like bacterial viruses and bacteriophages.
When a bacterium is infected by a bacteriophage, the bacterial virus inserts its DNA into the bacterial cell for the replication. The restriction enzyme produced within the bacterium halts the bacteriophage replication by cutting its DNA into several pieces. Restriction enzymes were named for their ability to restrict and provide a defense mechanism against the infection of viruses.
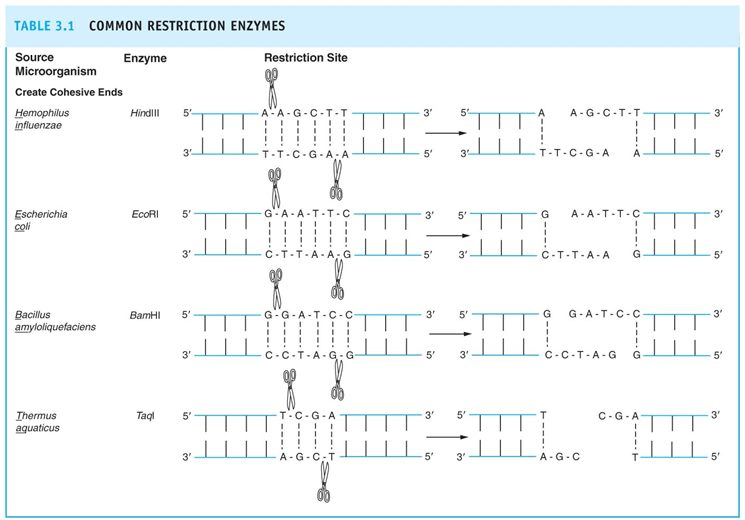
There are generally four types of restriction enzymes in which each enzyme recognizes a specific sequence of nucleotide bases (Adenine, Cytosine, Thymine and Guanine). These regions are randomly distributed in the DNA and are known as recognition sequences or recognition sites.
When restriction endonuclease enzyme recognizes a sequence of the DNA molecule, it catalyzes by hydrolysis i.e, addition of a water molecule which breaks the chemical bond between adjacent nucleotides. Methyltransferase enzyme adds methyl group (-CH3) to adenine and cytosine bases where bacterial DNA gets modified and protected from endonuclease enzymes. The restriction enzyme and its corresponding methyltransferase together form the restriction modification system.
Types of restriction enzymes
There are four types of restriction enzymes which are characterized on the basis of their structure, cleavage site, specificity and cofactor requirements.
- Type I enzyme
- Type II enzyme
- Type III enzyme
- Type IV enzyme
Type I enzyme:
- It is the first identified enzyme in two strains of E. coli (K-12 and B).
- Type I enzyme cuts at a site about 1000 bp away from the recognition site.
- The recognition site is composed of two specific portions: one with 3 to 4 nucleotides and other with 4 to 5 nucleotides which is separated by 6 to 8 nucleotides which is non specific and this site is asymmetrical.
- It posses both restriction digestion and modification methylase activities.
- Their cofactor includes adenosine triphosphate (ATP), S-Adenosyl methionine (AdoMet) and magnesium (Mg2+).
- Type I enzyme involves three subunits: HsdR for restriction digestion, HsdM for adding methyl groups (methyltransferase activity) and HsdS for recognition of DNA binding site (specificity).
- Some of the examples of Type I enzymes are EcoB and EcoK.
Type II enzyme:
- Type II enzyme cleave within a short specific distance of about 4 to 8 nucleotides in length and the sites are usually undivided.
- They cleave the phosphodiester bond of DNA either as a blunt end or sticky end.
- They do not use ATP or AdoMet but require Mg2+ as a cofactor.
- Type II enzyme is a homodimeric enzyme and are most commonly used enzymes.
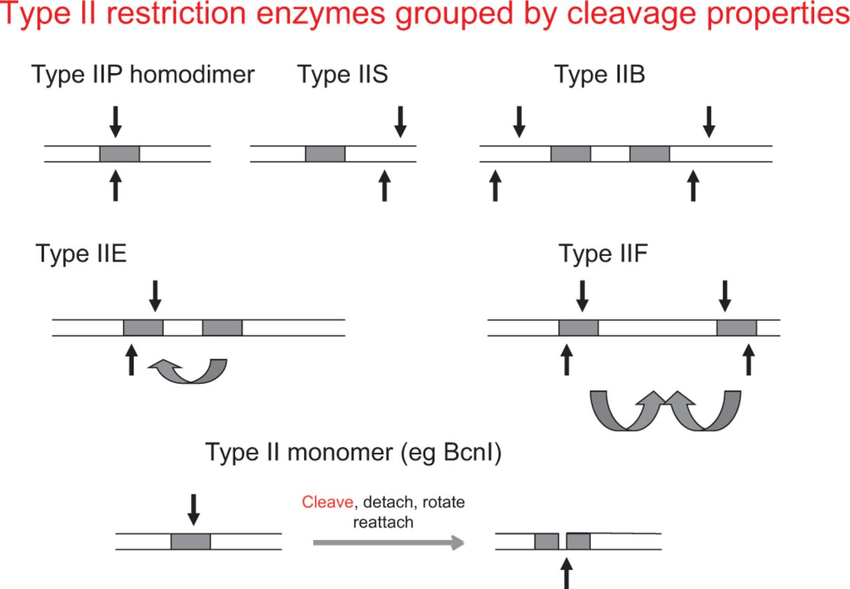
- There are seven subtypes of type II enzyme with different characteristics:
- Type IIS (Eg: Fok I)
- Type IIE (Eg: Nae I)
- Type IIF (Eg: NgoM IV)
- Type IIT (Eg: Bpu10 I)
- Type IIG (Eg: Eco 571)
- Type IIB (Eg: Bcg I)
- Type IIM (Eg: DPNI-RO)
Type III enzyme:
- Type III enzyme recognizes two separate non-palindromic sequences and cleaves DNA at the site of 25 to 27 bp from the recognition site.
- Type II enzymes are composed of two subunits: Res and Mod.
- The Res subunit is required for restriction digestion and the Mod subunit is for the recognition of DNA sequence and methyltransferase modification. Therefore, Type III enzymes are multifunctional and hetero-oligomeric.
- Type III enzyme requires ATP and Mg2+ cofactor for their role in DNA methylation and restriction digestion.
- Type III enzyme methylate only one strand of DNA at the N-6 position of adenosine which is enough to protect it against restriction digestion.
- Some of the examples of Type III enzymes are EcoP I and Hinf III.
Type IV enzyme:
- Type IV enzyme cleaves near or close to the recognition sequence.
- It targets methylated DNA, hydromethylated DNA and glucosyl hydromethylated DNA which are all modified types of DNA.
- Some of the examples of Type IV enzymes in the systems of E. coli are McrBC and Mrr.
- There are also other type of restriction enzyme that are Type V enzyme and artificial restriction enzyme.
- Type V enzyme uses RNA as a guide to target specific sequences that cleaves DNA of variable length. The cas9-gRNA complex from CRISPRs is an example of Type V restriction enzyme.
- Artificial restriction enzymes can be made by engineering DNA binding domain to a nuclease domain. Artificial restriction enzymes are used in genetic engineering and gene cloning. Zinc finger nuclease is an example of artificial restriction enzyme which is used commonly.
References:
- https://www.britannica.com/science/restriction-enzyme
- https://www.sigmaaldrich.com/technical-documents/protocols/biology/restriction-endonucleases-the-molecular-scissors.html
- https://www.nature.com/scitable/topicpage/restriction-enzymes-545/