The nucleotide salvage pathway is a metabolic mechanism in which organisms recycle nucleotides by recycling the purine and pyrimidine bases produced during RNA and DNA breakdown.
Nucleotides are the primary building blocks required for DNA and RNA synthesis.
Nucleotides are classified into two types: pyrimidines and purines. Each nucleotide is made up of three functional groups: a sugar, a base, and a phosphate. Purine and pyrimidine nucleotides are important energy transporters, nucleic acid subunits, and precursors for the synthesis of nucleotide cofactors like NAD and SAM.
The pyrimidine family contains thymine (T), cytosine (C), and uracil (U), which is exclusively found in RNA. A single-ringed nitrogenous base pairs with a purine nucleotide counterpart in these molecules. In contrast to cytosine, which couples with guanine to generate three hydrogen bonds, thymine partners with adenine to form two hydrogen bonds. Purines, including guanine (G) and adenine (A), are double-ringed molecules that are more resistant to degrade in the body.
There are two pathways for the synthesis of nucleotides, salvage and de novo.
Define Nucleotide salvage pathway
The nucleotide salvage pathway is a metabolic mechanism in which organisms recycle nucleotides by recycling the purine and pyrimidine bases produced during RNA and DNA breakdown.
Instead of creating the bases from scratch, the salvage process transforms them into nucleotides by incorporating ribose or deoxyribose sugars. The nucleotides synthesized can then be utilized to create fresh DNA and RNA molecules. The nucleotide salvage mechanism saves energy and resources by recycling preexisting nucleotides instead of generating them from scratch.
Steps of Nucleotide Salvage Pathway
The stages of the nucleotide salvage process vary based on the base being salvaged, but here’s a broad overview of the process:
- As an outcome of DNA and RNA degradation, or turnover, free purine or pyrimidine bases get released into the cytoplasm.
- Purine or pyrimidine bases are identified by specialized enzymes that catalyze their conversion into nucleosides by adding a ribose or deoxyribose sugar to them. Adenosine kinase, for example, transforms free adenine into adenosine.
- Nucleoside kinases phosphorylate the nucleosides, forming nucleotides, which are the components that make up blocks of DNA and RNA. Adenylate kinase, for example, phosphorylates adenosine to generate adenosine monophosphate (AMP).
- As needed, the newly synthesized nucleotides can be integrated into DNA or RNA synthesis pathways.
It is important to note that certain enzymes are required for each stage of the nucleotide salvage process, and the enzymes involved might differ depending on the nucleotide being saved.
PRPP synthesis
Phosphoribosylpyrophosphate (PRPP) is an essential intermediate/substrate in the synthesis of nucleotides, including purine and pyrimidine nucleotides. PRPP is made from ribose-5-phosphate, which is either through the pentose phosphate pathway or from nucleotide breakdown.
Here are the general steps of PRPP synthesis:
- The enzyme phosphoribosylpyrophosphate synthetase (PRPS) converts ribose-5-phosphate to 5-phosphoribosyl-1-pyrophosphate (PRPP).
- To generate PRPP, PRPS catalyzes the shift of a pyrophosphate group from ATP to the C1 position of ribose-5-phosphate.
- A supply of magnesium ions is required for the process, which is hindered by purine nucleotides and their analogs.
PRPP serves as a substrate for the addition of purine bases by the enzymes involved in de novo synthesis or the salvage pathway.
Note that PRPP is also involved in other biochemical pathways, such as the biosynthesis of histidine, tryptophan, and NAD. Deficiencies in PRPS can lead to disorders such as X-linked phosphoribosylpyrophosphate synthetase superactivity, which is characterized by gout, hyperuricemia, and neurological symptoms.
Regulation of PRPP synthase
Pi (inorganic phosphate) activates PRPP synthase, which is inhibited by the purine bases adenine and guanine.
Purine Synthesis: Salvage Pathway
The salvage pathway for purine synthesis requires the recycling of free bases of purines such as hypoxanthine and guanine to create purine nucleotides such as adenosine monophosphate (AMP) & guanosine monophosphate (GMP). The following are the general stages of the purine salvage pathway:
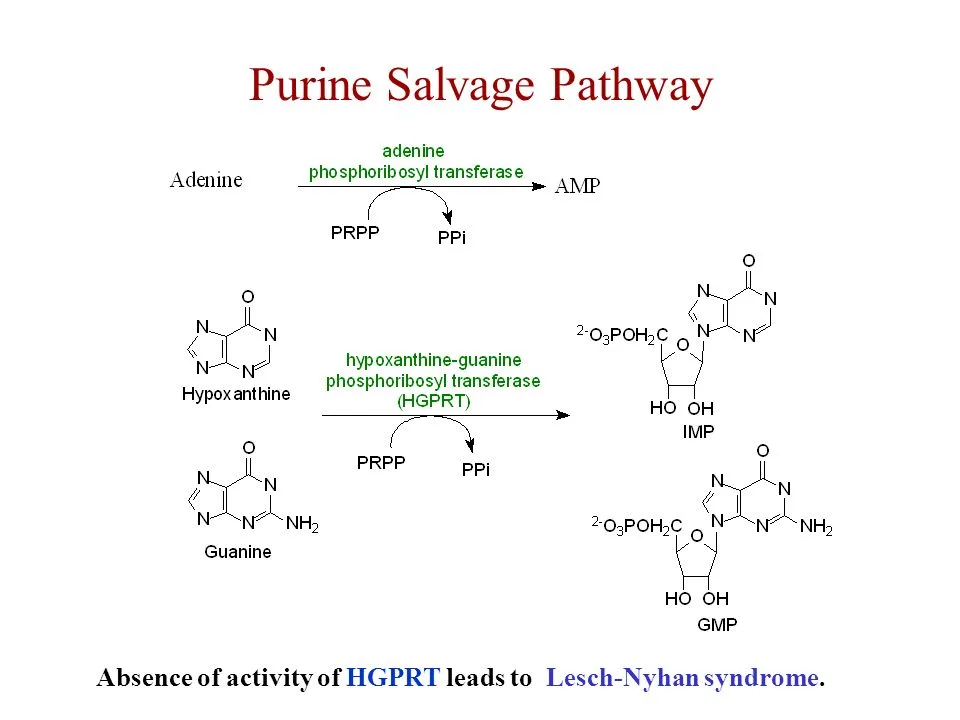
- Free purine bases, such as hypoxanthine and guanine, are released into the cytoplasm from degraded RNA and DNA.
- Hypoxanthine is converted to inosine monophosphate (IMP) by the enzyme hypoxanthine-guanine phosphoribosyltransferase (HGPRT), which catalyzes the transfer of a phosphoribosyl group from phosphoribosylpyrophosphate (PRPP) to hypoxanthine.
- Guanine is converted to guanosine monophosphate (GMP) by the enzyme guanine phosphoribosyltransferase (GPT), which also catalyzes the transfer of a phosphoribosyl group from PRPP to guanine.
- MP and GMP can then be further transformed to AMP and GMP, respectively, by a sequence of enzymatic processes including phosphate group transfer.
- The resultant purine nucleotides can be integrated as needed into DNA and RNA synthesis pathways.
It should be noted that the purine salvage route requires the employment of certain enzymes such as HGPRT and GPT, as well as the availability of PRPP as a substrate. Deficiencies in these enzymes can cause illnesses like Lesch-Nyhan syndrome and gout, which are characterized by a buildup of purine metabolites in the body.
Regulation of purine synthesis
PRPP allosterically activates the regulatory enzyme GPAT while inhibiting IMP, AMP, and GMP. To suppress the action of this enzyme, all three must be present.
Pyrimidine synthesis: Salvage pathway
The steps of the pyrimidine salvage pathway are as follows:
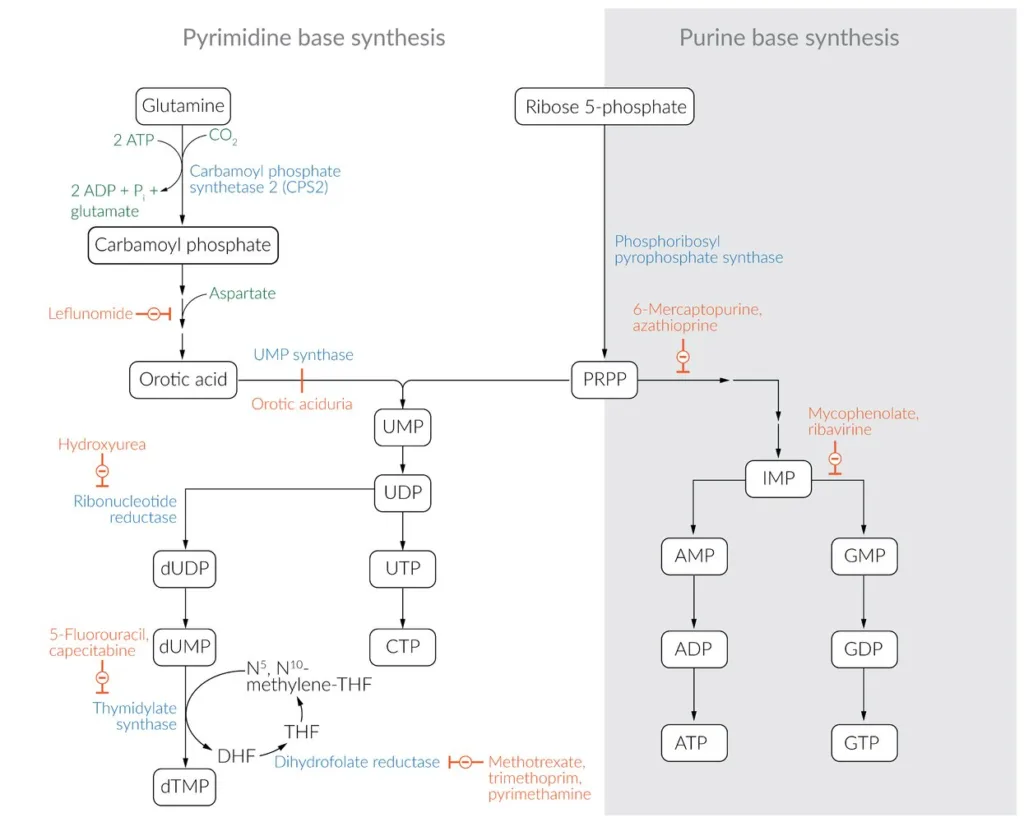
- Pyrimidine bases, such as uracil, thymine, and cytosine, can be recovered from degraded RNA and DNA or from dietary sources.
- Carbamoyl phosphate is synthesized from glutamine, carbon dioxide, and two ATP molecules by the enzyme carbamoyl phosphate synthetase II (CPSII).
- Aspartate is added to carbamoyl phosphate to form carbamoyl aspartate by the enzyme aspartate transcarbamoylase.
- Carbamoyl aspartate is then converted to dihydroorotate by the enzyme dihydroorotase.
- Dihydroorotate is oxidized to orotate by the enzyme dihydroorotate dehydrogenase.
- Orotate is then converted to orotidine-5′-monophosphate (OMP) by the enzyme orotate phosphoribosyltransferase (OPRT).
- OMP is then decarboxylated to uridine-5′-monophosphate (UMP) by the enzyme OMP decarboxylase.
- UMP can then be phosphorylated to form uridine-5′-diphosphate (UDP) and uridine-5′-triphosphate (UTP) by the action of nucleoside diphosphate kinase and nucleoside triphosphate kinase, respectively.
- UTP can be converted to cytidine triphosphate (CTP) by the action of CTP synthase.
- Alternatively, UMP can be converted to deoxyuridine-5′-monophosphate (dUMP) and further to deoxythymidine-5′-monophosphate (dTMP) by the action of thymidylate synthase. This reaction requires folate as a cofactor to provide a methyl group for the conversion of dUMP to dTMP.
The salvage pathway allows the cell to recycle pre-existing pyrimidine bases and nucleosides rather than having to synthesize them de novo from scratch, thereby conserving resources.
Regulation of pyrimidine synthesis
The reaction catalyzed by CSPII is the pathway’s regulatory step, which is triggered by PRPP and ATP and blocked by UTP.
Differences between the purine and pyrimidine salvage pathways
Here is a comparison table highlighting some of the key differences between the purine and pyrimidine salvage pathways:
| Feature | Purine Salvage Pathway | Pyrimidine Salvage Pathway |
| Precursor | PRPP | Carbamoyl phosphate |
| Base | Added onto ribose sugar | Synthesized first, then added onto ribose sugar |
| Enzyme | HGPRT, APRT | OPRT, OMP decarboxylase |
| End product | Purine nucleotide | Pyrimidine nucleotide |
| Key Intermediate | IMP | UMP |
| Conversion to other nucleotides | IMP can be converted to AMP and GMP | UMP can be converted to CTP, and can also be converted to dUMP for thymidine synthesis |
While both pathways involve the recycling of nucleotide components, the specific precursors, order of synthesis, enzymes, and end products are different.