What is RFLP?
Restriction Fragment Length Polymorphism (RFLP) is a molecular biology technique that examines DNA sequence variations using specific enzymes known as restriction enzymes. These enzymes identify and cut DNA at specific sites called restriction sites, generating smaller fragments. Gel electrophoresis is then used to separate the DNA fragments, enabling their visualization and analysis. These variations in fragment sizes, referred to as polymorphisms, provide valuable information about genetic distinctions among individuals or species. RFLP has been extensively applied in genetic mapping, exploring genetic diversity, and forensic identification. However, it has been largely surpassed by more advanced DNA sequencing methods. Nonetheless, RFLP remains an essential tool in genetic research due to its historical significance and ability to provide valuable genetic insights.
Principle of RFLP
The fundamental principle of Restriction Fragment Length Polymorphism (RFLP) is founded on the ability of specific enzymes called restriction enzymes or restriction endonucleases to divide DNA molecules into smaller fragments. These enzymes have the capability to identify particular DNA sequences, referred to as restriction sites, and cleave the DNA at or in close proximity to these sites. As a consequence of this cleavage, the resulting DNA fragments exhibit variations in size, generating a distinctive fragment pattern that is unique to each individual or species.
How RFLP analysis works?
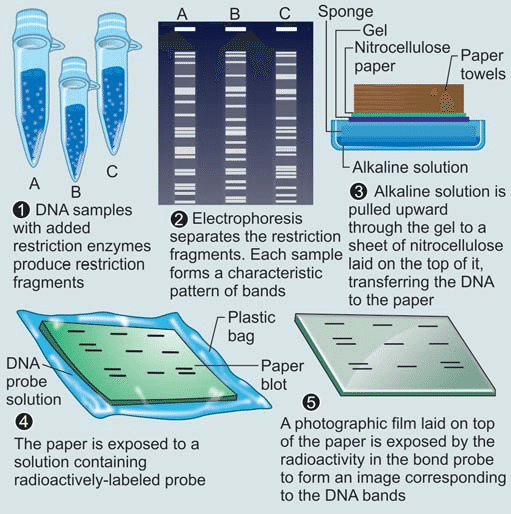
RFLP analysis involves several key steps to analyze and compare variations in DNA sequences. Here is a step-by-step explanation of how RFLP analysis works:
1. DNA Extraction:
DNA is extracted from the cells of the organism or sample of interest. This typically involves breaking open the cells and isolating the DNA using specialized extraction protocols.
2. Restriction Digestion:
The extracted DNA is treated with specific restriction enzymes (also known as restriction endonucleases). These enzymes recognize specific DNA sequences, known as restriction sites, and cleave the DNA at or near these sites. The choice of restriction enzymes depends on the target DNA sequence and the desired outcome of the analysis.
3. Gel Electrophoresis:
The DNA fragments obtained from the restriction digestion are separated based on their size using a technique called gel electrophoresis. In this process, a gel matrix, often made of agarose or polyacrylamide, is prepared and loaded into a gel tray. Small wells are created at one end of the gel, and the DNA samples, along with a size marker, are loaded into these wells.
4. Applying an Electric Field:
Once the DNA samples are loaded, an electric field is applied across the gel by immersing the gel tray in a buffer solution and connecting it to a power supply. The electric field causes the negatively charged DNA fragments to migrate through the gel matrix towards the positive electrode.
5. Separation by Size:
During gel electrophoresis, the DNA fragments move through the gel at different rates based on their size. Smaller fragments migrate more quickly through the gel, while larger fragments move more slowly. This separation by size allows for the visualization and analysis of the DNA fragments.
6. DNA Visualization:
After the electrophoresis run, the DNA fragments are visualized using staining techniques or other detection methods. Common staining methods involve treating the gel with DNA-specific dyes that bind to the DNA fragments and make them visible under ultraviolet (UV) light. Alternatively, radioactive or fluorescent labels can be used for detection, depending on the specific experimental setup.
7. Analysis of Fragment Patterns:
The resulting pattern of DNA fragments on the gel, often referred to as a “restriction pattern,” is analyzed and compared among different individuals or samples. The variations in fragment sizes, known as polymorphisms, indicate genetic differences. The restriction patterns can be recorded and interpreted, allowing for the identification of specific alleles or genetic markers associated with particular DNA sequences.
RFLP analysis has been widely used in various fields, including genetic mapping, forensic analysis, population genetics, and disease studies. However, it has been largely replaced by more advanced techniques such as polymerase chain reaction (PCR) and DNA sequencing, which offer greater speed, sensitivity, and specificity.
RFLP-PCR
RFLP-PCR (Restriction Fragment Length Polymorphism-Polymerase Chain Reaction) is a modified approach that combines the principles of RFLP analysis and PCR amplification. This technique overcomes some of the limitations of traditional RFLP analysis, such as the need for a large amount of high-quality DNA and the limited resolution.
The RFLP-PCR technique involves the following steps:
1. DNA Extraction:
Similar to traditional RFLP analysis, DNA is extracted from the cells of interest using standard methods.
2. PCR Amplification:
Instead of directly subjecting the extracted DNA to restriction digestion, PCR amplification is performed to selectively amplify the target DNA region. PCR involves the use of specific primers that flank the DNA sequence of interest. The primers bind to the DNA template, and through repeated cycles of heating and cooling, multiple copies of the target DNA region are synthesized.
3. Restriction Digestion:
After PCR amplification, the amplified DNA products, known as amplicons, are subjected to restriction digestion. Restriction enzymes, which recognize specific DNA sequences, are used to cleave the amplified DNA at or near their recognition sites. This generates DNA fragments with different lengths based on the presence or absence of restriction sites within the target DNA sequence.
4. Gel Electrophoresis and Analysis:
The resulting DNA fragments from the restriction digestion are separated by gel electrophoresis, similar to traditional RFLP analysis. The fragments are visualized and analyzed to determine the pattern of DNA banding. The fragment sizes can be compared among individuals or samples to identify genetic variations and polymorphisms.
RFLP-PCR offers several advantages over traditional RFLP analysis. It allows for the amplification of limited or degraded DNA samples, as PCR can generate sufficient DNA material for analysis. The technique also provides increased sensitivity, as PCR amplification can detect smaller amounts of DNA. Additionally, RFLP-PCR allows for the multiplexing of multiple target DNA regions in a single reaction, enabling the simultaneous analysis of multiple genetic markers.
However, it is important to note that RFLP-PCR still requires careful optimization and validation, and it may not offer the same level of resolution as other modern molecular techniques like DNA sequencing or SNP genotyping arrays. The choice of restriction enzymes and primer design in PCR amplification plays a critical role in the success and accuracy of RFLP-PCR analysis.
Overall, RFLP-PCR combines the advantages of PCR amplification and RFLP analysis, providing an enhanced approach for genetic variation analysis in various fields such as population genetics, forensic science, and disease studies.
Applications of RFLP
Restriction Fragment Length Polymorphism (RFLP) analysis has been widely used in various applications within the field of molecular biology. Some of the key applications of RFLP are:
Genetic Mapping:
RFLP analysis has played a crucial role in genetic mapping studies, particularly in the early stages of genome mapping. By identifying and analyzing DNA polymorphisms, RFLP markers were used to construct genetic linkage maps and locate genes of interest on specific chromosomes.
Population Genetics:
It has been employed to study genetic diversity and population structure. By examining the distribution of RFLP alleles within and between populations, scientists can gain insights into evolutionary relationships, migration patterns, and genetic differentiation among different groups.
Forensic Science:
RFLP analysis has been used in forensic investigations for DNA profiling and individual identification. By comparing RFLP patterns from crime scene samples with those of potential suspects, forensic scientists can assess genetic matches or exclusions, providing valuable evidence in criminal investigations.
Paternity Testing:
RFLP analysis has been utilized in determining biological parentage by comparing the DNA profiles of an individual and potential parents. RFLP markers can help establish genetic relatedness and resolve paternity disputes.
Disease Studies:
RFLP analysis has been applied in the study of genetic diseases and their inheritance patterns. By examining RFLP markers linked to disease-associated genes, researchers can investigate the presence or absence of specific genetic variants in affected individuals and their families.
Evolutionary Biology:
Restriction Fragment Length Polymorphism has contributed to the understanding of genetic variation and evolutionary processes. By comparing RFLP patterns among different species or populations, scientists can explore genetic relationships, identify evolutionary lineages, and study the mechanisms of speciation.
Plant and Animal Breeding:
RFLP analysis has been used in agricultural and animal breeding programs to assess genetic variation and make informed breeding decisions. By selecting individuals with desired RFLP patterns associated with favorable traits, breeders can improve the genetic composition of crops or livestock.
Advantages and Disadvantages of RFLP
RFLP (Restriction Fragment Length Polymorphism) analysis has several advantages and disadvantages, which are important to consider when choosing a molecular biology technique. Here are the key advantages and disadvantages of RFLP:
Advantages of RFLP:
- Genetic Variation Analysis: Restriction Fragment Length Polymorphism analysis allows the detection and analysis of genetic variations within and between individuals or species. It provides information on the presence or absence of specific DNA fragments, enabling the study of genetic diversity and relationships.
- Established Technique: RFLP has a long history and has been extensively used in genetic research. As a well-established technique, there is a wealth of literature and resources available for its implementation and interpretation.
- Genome Mapping: RFLP markers have been used for genetic mapping, aiding in the construction of genetic linkage maps and the localization of genes of interest to specific chromosomal regions.
- Historical Significance: RFLP analysis played a significant role in early genetic studies, contributing to our understanding of genetic variation, inheritance patterns, and evolutionary processes.
Disadvantages of RFLP:
- Labor-Intensive and Time-Consuming: RFLP analysis involves multiple steps, including DNA extraction, restriction digestion, gel electrophoresis, and DNA visualization. The process can be time-consuming and requires technical expertise, making it less efficient compared to newer molecular techniques.
- Limited Resolution: RFLP analysis has limited resolution in detecting small-scale genetic variations. It primarily detects differences in the length of DNA fragments, and the resolution is dependent on the restriction enzymes used and the number of restriction sites within the DNA sequence of interest.
- Low Sensitivity: RFLP analysis may have lower sensitivity compared to other techniques like PCR-based methods. It requires a relatively large amount of high-quality DNA for analysis, which may pose challenges when dealing with limited or degraded DNA samples.
- Lack of Multiplexing: It typically focuses on a single DNA fragment or a limited number of fragments at a time. Multiplexing, the simultaneous analysis of multiple DNA markers, is more challenging with RFLP compared to PCR-based methods.
- Reduced Applicability: With the advent of more advanced molecular techniques such as PCR and DNA sequencing, the application of RFLP has diminished over time. These newer methods offer higher sensitivity, specificity, and flexibility, making them more suitable for many research and diagnostic purposes.
References:
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., & Walter, P. (2002). Molecular Biology of the Cell. Garland Science.
- Brown, T. A. (2017). Gene Cloning and DNA Analysis: An Introduction. Wiley-Blackwell.
- Hartl, D. L., & Jones, E. W. (2005). Genetics: Analysis of Genes and Genomes. Jones & Bartlett Learning.
- Lodish, H., Berk, A., Zipursky, S. L., Matsudaira, P., Baltimore, D., & Darnell, J. (2000). Molecular Cell Biology. W. H. Freeman.
- Russell, P. J., Hertz, P. E., & McMillan, B. (2010). Biology: The Dynamic Science. Cengage Learning.