Discover the intricate world of purine and pyrimidine biosynthesis through the de novo pathway. These essential nucleotides are fundamental building blocks for DNA and RNA synthesis, playing vital roles in various cellular processes. Uncover the steps, energy calculations, applications, limitations, and inhibitory factors associated with De Novo Purine and Pyrimidine Biosynthesis and Regulation to gain a comprehensive understanding of their significance in cellular metabolism.
Steps of De Novo Purine Biosynthesis:
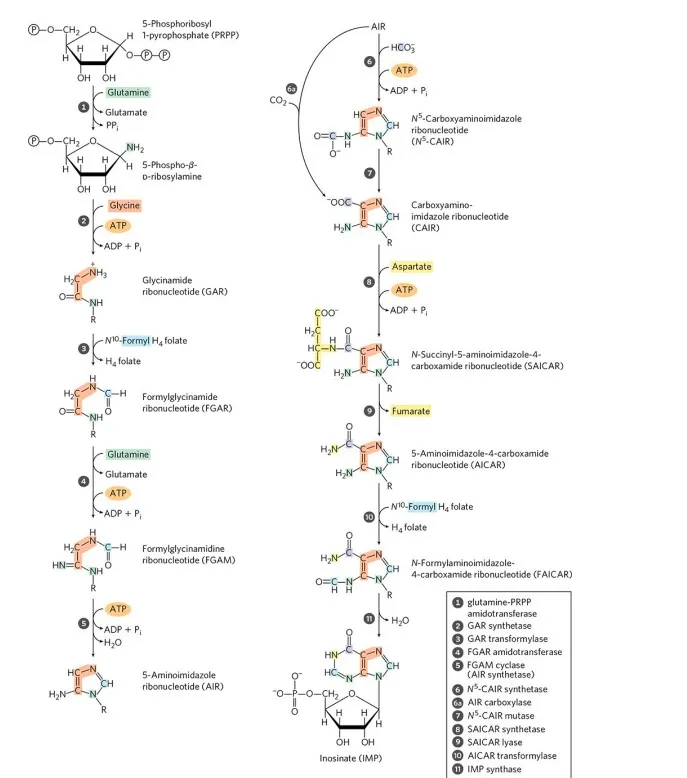
Embark on the journey of purine biosynthesis as it unfolds step by step:
- Ribose-5-Phosphate Conversion: Ribose-5-phosphate, derived from the pentose phosphate pathway, is transformed into phosphoribosyl pyrophosphate (PRPP) by the enzyme PRPP synthetase, utilizing ATP.
- Formation of 5-Phosphoribosylamine: PRPP combines with glutamine under the guidance of the enzyme amidophosphoribosyltransferase (PPAT), leading to the formation of 5-phosphoribosylamine, a critical intermediate in purine biosynthesis.
- Purine Ring Assembly: The purine ring takes shape on the 5-phosphoribosylamine scaffold through a series of enzymatic reactions, gradually forming inosine monophosphate (IMP), a purine nucleotide.
- Conversion to Adenosine and Guanosine: IMP can undergo further enzymatic transformations to generate adenosine monophosphate (AMP) or guanosine monophosphate (GMP). Aspartate contributes an amino group for the conversion to AMP, while glutamine provides the necessary amino group for GMP synthesis.
- Phosphorylation to ATP and GTP: AMP and GMP can be phosphorylated to yield adenosine triphosphate (ATP) and guanosine triphosphate (GTP), respectively. These high-energy molecules play essential roles in cellular energy metabolism and signaling processes.
Steps of De Novo Pyrimidine Biosynthesis:
Uncover the sequential enzymatic transformations that shape pyrimidine biosynthesis through the de novo pathway:
- Carbamoyl Phosphate Formation: Bicarbonate and glutamine join forces in the presence of ATP, catalyzed by carbamoyl phosphate synthetase II (CPS II), to produce carbamoyl phosphate.
- Formation of Orotate: Carbamoyl phosphate combines with aspartate under the guidance of dihydroorotase, forming dihydroorotate. This compound is subsequently oxidized to orotate by dihydroorotate dehydrogenase.
- Activation of Orotate: The addition of phosphoribosyl pyrophosphate (PRPP) to orotate, facilitated by orotate phosphoribosyltransferase (OPRTase), results in the formation of orotidine 5′-monophosphate (OMP).
- Conversion to UMP and CMP: OMP undergoes decarboxylation via orotidine-5′-phosphate decarboxylase (OMP decarboxylase), producing uridine 5′-monophosphate (UMP). Alternatively, cytidylate synthetase orchestrates a series of enzymatic reactions, leading to the formation of cytidine 5′-monophosphate (CMP).
- Conversion to UTP and CTP: UMP and CMP are further phosphorylated, generating uridine triphosphate (UTP) and cytidine triphosphate (CTP), respectively. These nucleotides serve crucial functions in cellular processes and signaling pathways.
Regulation Mechanism of De Novo Purine and Pyrimidine Biosynthesis:
Delve into the intricate regulatory mechanisms that govern de novo purine and pyrimidine biosynthesis pathways:
- Feedback Inhibition: AMP, GMP, ADP, GDP (for purines), and UTP, CTP (for pyrimidines) act as feedback inhibitors, allosterically inhibiting specific enzymes in earlier steps of their respective pathways. This mechanism ensures balanced nucleotide pools by reducing pathway activity when nucleotide levels are sufficient.
- Enzyme Regulation: Key enzymes in the pathways are subject to regulation through allosteric modulation and covalent modifications like phosphorylation. For example, PRPP activates glutamine phosphoribosyl pyrophosphate amidotransferase (GPAT) in purine biosynthesis, while AMP and GMP inhibit it. In pyrimidine biosynthesis, ATP and UTP regulate carbamoyl phosphate synthetase II (CPS II).
- Gene Expression Control: Transcription factors and regulatory proteins influence the expression of genes encoding enzymes involved in de novo biosynthesis. These proteins bind to specific DNA sequences in gene promoters, controlling their transcription. Substrate or end product availability can impact gene expression, fine-tuning pathway enzyme levels.
- Hormonal Regulation: Hormones, such as insulin and glucagon, play a role in regulating nucleotide biosynthesis. Insulin stimulates synthesis, while glucagon inhibits it. Hormonal signaling pathways modulate enzyme activity or gene expression, influencing nucleotide production.
- Cellular Signaling and Metabolic Status: Signaling pathways like mTOR and AMP-activated protein kinase (AMPK) integrate cellular nutrient and energy status to regulate biosynthesis. These pathways monitor metabolite levels and adjust enzyme activity accordingly.
Energy Calculations:
The de novo biosynthesis of purines and pyrimidines demands the utilization of ATP and GTP as energy sources. The conversion of ribose-5-phosphate to PRPP consumes one ATP molecule, and subsequent steps involve additional ATP and GTP molecules, making the process energy-intensive.
Applications and Limitations:
Purine and pyrimidine nucleotides synthesized via the de novo pathway find extensive applications in DNA and RNA replication, transcription, translation, and cellular signaling. Additionally, they serve as precursors for coenzyme synthesis (e.g., NAD+, FAD) and secondary messenger production (e.g., cAMP, cGMP).
However, the de novo pathway has limitations. Genetic defects, enzyme deficiencies, or metabolic disorders can disrupt the pathway due to its dependence on various enzymes and cofactors. Moreover, excessive purine and pyrimidine nucleotide levels can lead to metabolic imbalances like hyperuricemia or gout.
Inhibition of De Novo Purine and Pyrimidine Biosynthesis:
Certain compounds act as inhibitors, targeting specific enzymes in the de novo pathways. Methotrexate, for example, inhibits dihydrofolate reductase, a crucial enzyme involved in tetrahydrofolate synthesis required for nucleotide biosynthesis.
Differences between the purine and pyrimidine de novo pathways
| Parameter | Purine De Novo Pathway | Pyrimidine De Novo Pathway |
|---|---|---|
| Precursors | Ribose-5-phosphate | Bicarbonate, aspartate, and glutamine |
| Key Intermediates | Phosphoribosyl pyrophosphate (PRPP) | Orotidine 5′-monophosphate (OMP) |
| Ring Formation | Purine ring assembly | Pyrimidine ring formation |
| Energy Usage | ATP and GTP | ATP |
| Enzymes | PRPP synthetase, amidophosphoribosyltransferase | Carbamoyl phosphate synthetase II, orotate phosphoribosyltransferase |
| End Products | AMP, GMP | UMP, CMP |
| Final Products | ATP, GTP | UTP, CTP |
References:
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., & Walter, P. (2014). Molecular Biology of the Cell (6th ed.). Garland Science.
- Nelson, D. L., Cox, M. M. (2017). Lehninger Principles of Biochemistry (7th ed.). W. H. Freeman and Company.
- Berg, J. M., Tymoczko, J. L., & Gatto, G. J. (2018). Stryer’s Biochemistry (8th ed.). W.H. Freeman and Company.
Learn more: