In the field of molecular biology, the ability to accurately quantify nucleic acids is fundamental to numerous applications, spanning from basic research to clinical diagnostics. While traditional quantitative PCR (qPCR) has been the most useful technique for nucleic acid quantification, recent advancements have led to the emergence of a groundbreaking technology: digital droplet polymerase chain reaction (ddPCR). Offering unparalleled precision, sensitivity, and absolute quantification capabilities, ddPCR has revolutionized the landscape of nucleic acid analysis.
Digital droplet PCR (ddPCR) represents a groundbreaking advancement in nucleic acid quantification, offering unparalleled precision, sensitivity, and absolute quantification capabilities. At the heart of ddPCR lies sophisticated instrumentation designed to partition samples into discrete droplets, enabling precise quantification of target DNA or RNA molecules.
What is ddPCR?
Digital droplet PCR (ddPCR) stands as a pinnacle of innovation in nucleic acid quantification. At its core, ddPCR is an advanced iteration of traditional PCR, employing the novel strategy of partitioning a sample into thousands of minute droplets, each acting as an independent reaction chamber. This partitioning allows for absolute quantification of target DNA or RNA molecules, which is unattainable through conventional PCR methods.
Steps of ddPCR
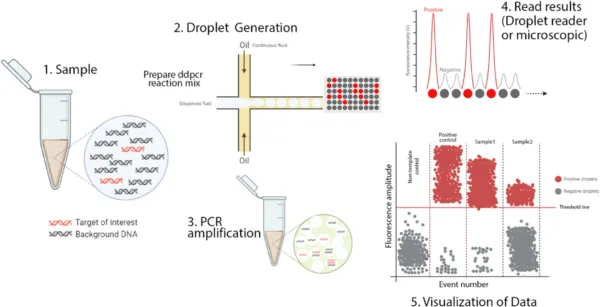
The ddPCR process works through the following steps:
Sample Partitioning:
The sample undergoes partitioning using water-oil emulsion or microfluidics, generating numerous individual reactions within distinct droplets.
Amplification:
Within each droplet, PCR amplification proceeds, facilitated by the inclusion of PCR reagents such as primers, nucleotides, and DNA polymerase.
Endpoint Analysis:
Following amplification, droplets are scrutinized through fluorescence detection methods to ascertain the presence or absence of amplified target sequences.
Digital Quantification:
Crucially, dd PCR achieves digital quantification by counting the number of positive and negative droplets, enabling precise determination of target molecule concentrations in the original sample.
Data Analysis:
The acquired fluorescence data is meticulously analyzed to calculate absolute target nucleic acid concentrations, a hallmark of ddPCR’s accuracy and reliability.
Discovery and Evolution of ddPCR
The genesis of droplet PCR can be traced back to the pioneering work of researchers at Raindance Technologies and the National Institute of Standards and Technology (NIST), USA. In the early 2000s, Dr. Fred Kramer and his team embarked on a journey to conceptualize and refine the principles of partitioned PCR, laying the groundwork for ddPCR’s eventual realization.
A seminal moment in ddPCR’s trajectory occurred with the publication of Hindson et al.’s seminal paper in Analytical Chemistry in 2011. This landmark study elucidated the microfluidic emulsion technology pivotal for partitioning samples into droplets, marking a watershed moment in the evolution of ddPCR. Subsequent efforts by Raindance Technologies propelled ddPCR into the scientific limelight, facilitating its widespread adoption in research laboratories worldwide.
Principles of ddPCR
The principles underpinning ddPCR encapsulate a fusion of traditional PCR methodologies with innovative digital quantification capabilities. The meticulous partitioning of samples into discrete droplets serves as the cornerstone of ddPCR’s precision and accuracy. By enabling absolute quantification without reliance on standard curves, ddPCR transcends the limitations of traditional qPCR, offering researchers unparalleled insights into nucleic acid dynamics.
Advantages of ddPCR
The advantages conferred by ddPCR over traditional qPCR methodologies are manifold:
- Improved Precision: ddPCR heralds a new era of precision in nucleic acid quantification, particularly at low concentrations, mitigating variability and enhancing accuracy.
- Enhanced Sensitivity: ddPCR’s heightened sensitivity empowers researchers to detect and quantify minute quantities of target molecules, opening vistas for rare mutation detection and viral nucleic acid analysis.
- Resistance to PCR Inhibitors: ddPCR’s resilience to PCR inhibitors underscores its utility in analyzing complex or impure samples, ensuring reliable quantification even in challenging experimental contexts.
- Absolute Quantification: Unlike qPCR’s reliance on relative quantification, droplet digital PCR facilitates absolute quantification, obviating the need for external calibration and enhancing result robustness.
- Reduced Variability: By virtue of partitioning samples into individual droplets, ddPCR mitigates variability stemming from pipetting errors and sample-to-sample variations, engendering reproducible and reliable data.
- Multiplexing Capability: ddPCR’s multiplexing prowess empowers researchers to simultaneously quantify multiple targets within a single sample, streamlining assay workflows and conserving resources.
- Applications in Liquid Biopsy and Clinical Diagnostics: ddPCR’s unparalleled sensitivity renders it indispensable in liquid biopsy and clinical diagnostics, facilitating the detection of circulating tumor DNA and genetic mutations with unprecedented accuracy.
Limitations of ddPCR
However, despite its transformative potential, ddPCR is not devoid of limitations:
- Cost: The initial capital investment and per-sample costs associated with ddPCR instrumentation and consumables may pose barriers to adoption, particularly for resource-constrained laboratories.
- Throughput: ddPCR systems often exhibit lower throughput compared to traditional qPCR platforms, limiting their scalability for high-throughput applications.
- Complex Sample Preparation: The intricacies of droplet generation and sample partitioning in ddPCR necessitate meticulous attention to experimental protocols, potentially impeding workflow efficiency.
- Limited Dynamic Range: ddPCR’s dynamic range may be constrained, requiring dilution or concentration steps for accurate quantification of target molecules falling outside the linear range of detection.
- Assay Design Challenges: Designing optimized assays necessitates careful consideration of factors such as droplet stability and PCR conditions, demanding expertise and iterative optimization.
- Data Interpretation Complexity: Interpreting data mandates sophisticated analytical tools and computational expertise, particularly for nuanced data sets or complex experimental designs.
- Multiplexing Constraints: While ddPCR enables multiplexing, the multiplexing capacity may be restricted compared to qPCR, necessitating judicious assay design and optimization.
Conclusion
In conclusion, digital droplet PCR stands as a paragon of innovation in nucleic acid quantification, offering unprecedented precision, sensitivity, and absolute quantification capabilities. From its inception to its widespread adoption across diverse research domains, it exemplifies the power of interdisciplinary collaboration and technological ingenuity in driving scientific progress. While acknowledging its limitations, the transformative potential of ddPCR in elucidating the intricacies of nucleic acid dynamics remains unequivocal, paving the way for a new era of precision molecular biology and clinical diagnostics.