DNA microarray technique is a tool for the analysis of the expression of many thousands of genes at once. The area of genomics has undergone a revolution as a result, offering fresh perspectives on the underlying genetic mechanisms of disease and creating new opportunities for the advancement of customized medicine. We will study about the underlying concepts, uses, and effects of DNA microarray technology across a range of scientific disciplines in this article.
What is DNA Microarray Technology?
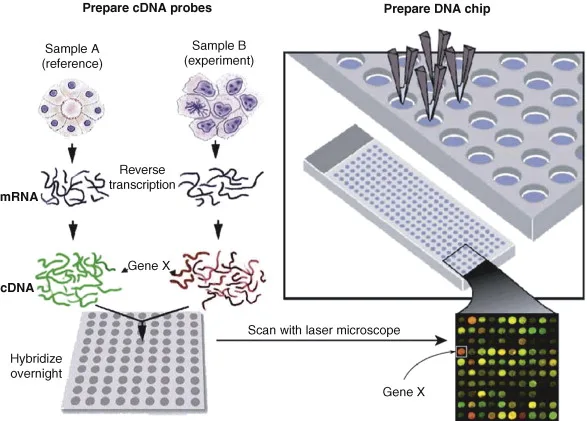
DNA microarray technology is a high-throughput method for examining how genes are expressed. It requires affixing tens of thousands of DNA probes, each of which corresponds to a distinct gene, to a solid, flat surface, like a glass slide or membrane. The probes are made to bind or hybridize with complementary DNA sequences that have been taken from biological samples like cells or tissues.
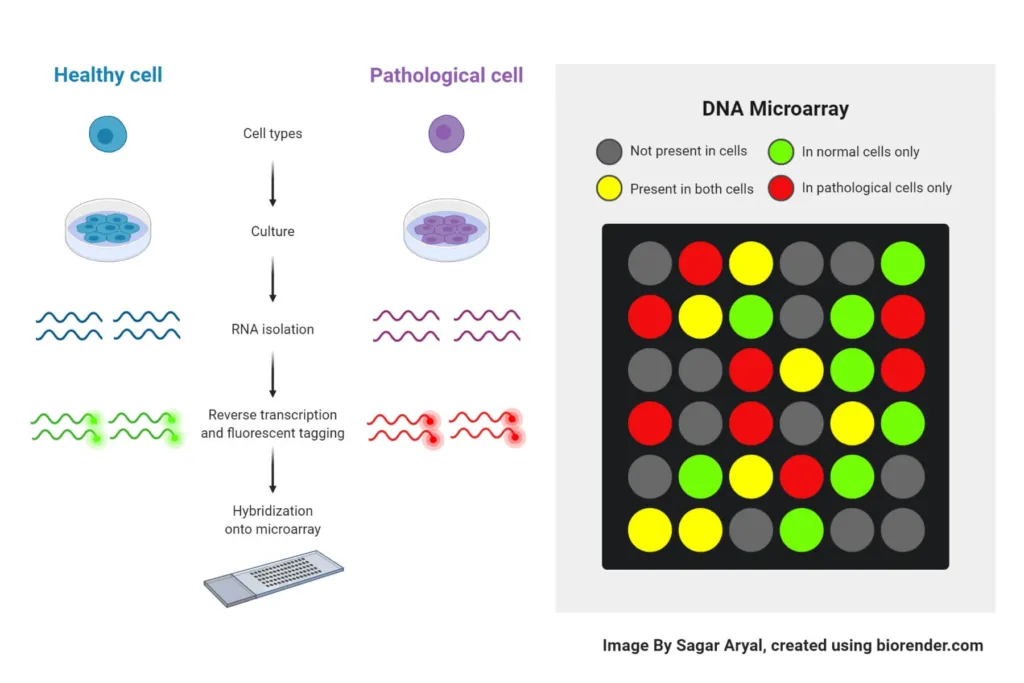
The biological material is tagged with one fluorescent dye, and the DNA probes are labeled with another fluorescent dye to study gene expression. After that, the sample and probes are mixed together and given time to hybridize. Using a scanner or another imaging instrument, the resultant fluorescent signal is identified and examined. Each probe’s signal intensity reflects how much of the gene it represents is expressed in the biological sample.
Principles of DNA Microarray technology
- The Principle of DNA microarray technology is the capacity of complementary DNA strands to hybridize, or bond, with one another. A biological sample’s complementary DNA sequences are intended to hybridize with the little pieces of DNA, or probes, that make up a DNA microarray.
- The joining of two complementary DNA strands by hydrogen bonding between the nucleotide base pairs is the process of DNA hybridization. DNA is composed of the nucleotide bases adenine (A), thymine (T), cytosine (C), and guanine (G). Adenine always pairs with thymine, and cytosine always pairs with guanine, to form base pairs.
- Typically, small DNA segments are attached to a glass slide or membrane to create the DNA microarray probes. These probes include fluorescent tags that make them detectable and are designed to complement certain genes of interest.
- In order to conduct a DNA microarray experiment, the biological material must first be processed by removing the DNA and utilizing reverse transcription to transform it into complementary RNA (cRNA). Then, to enable detection and comparison, the cRNA is labeled with an alternative fluorescent tag, often one that is of a different color than the probe.
- After that, the labeled cRNA is put to the microarray, where it hybridizes with the corresponding cRNA sequences in the sample using the probes as a guide. The fluorescent signal from each probe, which represents the degree of the matching gene’s expression in the sample, is then detected by scanning the microarray.
- The expression levels of thousands of genes may be at once determined by examining the fluorescence signal from each probe. Insights into the molecular mechanisms behind these disorders are gained by being able to identify genes that are upregulated or downregulated in particular circumstances, such as illness or pharmacological therapy.
Instrumentation of DNA Microarray technique
Several types of equipment are needed for the DNA microarray approach, and they are employed at various phases of the process. Here are some of the major instruments used in DNA microarray technique:
- Microarray printer: DNA probes are created and printed onto a glass slide or another solid surface using a microarray printer. A pattern of probes that can be used to measure gene expression levels may be created by programming the printer to drop small droplets of DNA solution onto the slide.
- Hybridization oven: The microarray slide and the labeled sample are incubated together in a hybridization oven. The hybridization of the probes and sample is made possible by the oven’s regulated temperature and humidity conditions.
- Microarray scanner: The fluorescent signals produced by the hybridized probes and labeled material are picked up by a microarray scanner. A quantitative measurement of the levels of gene expression may be obtained from the scanner by detecting the fluorescence intensity that each probe on the slide emits.
- Data analysis software: The microarray scanner’s data is analyzed using specialized software. The program can provide graphical representations of gene expression patterns, normalize the data, and remove noise.
- Real-time PCR machine: The outcomes of a DNA microarray study can be verified using a real-time PCR equipment. Real-time PCR is able to measure the sample’s expression levels of certain genes and validate the outcomes of microarray analysis.
Other equipment that may be used in DNA microarray technique includes a microarray washing station, pipettes, centrifuges, and spectrophotometers for measuring the concentration and purity of DNA and RNA samples. The specific instrumentation required for DNA microarray technique can vary depending on the experimental design and the platform used.
Steps of DNA Microarray analysis
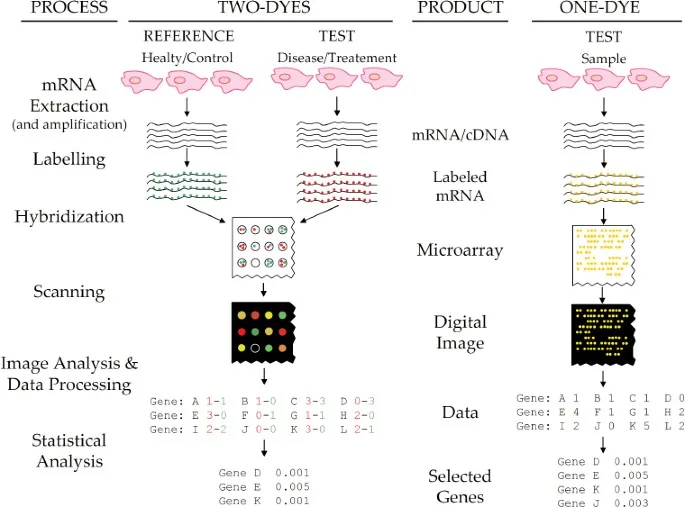
The procedure for DNA microarray technique involves several steps, including sample preparation, probe design and synthesis, labeling and hybridization, and data analysis. Here is a general overview of the procedure:
- Sample preparation: The preparation of the biological sample for DNA microarray analysis is the first stage in the process. Depending on the experimental setup, this may include separating DNA or RNA from cells or tissues. Accurate findings depend on the quality and size of the sample.
- Probe design and synthesis: Following sample preparation, probes are made to hybridize with certain target genes. A number of techniques, including photolithography and inkjet printing, can be used to create probes. Typically, a glass slide or membrane is used as the solid surface on which the probes are fastened.
- Labeling and hybridization: To facilitate detection and comparison, the fluorescent tag on the biological sample is often a distinct color from the probes. Following application of the labeled sample to the microarray, hybridization between the probes and the corresponding sequences in the sample takes place. After then, the microarray is washed to get rid of any unbound samples.
- Scanning and data analysis: The fluorescent signal from each probe, which represents the degree of the matching gene’s expression in the sample, is detected by scanning the microarray. A gene expression profile is then created by quantifying and analyzing the fluorescence signal using specialist software.
- Validation: Other experimental techniques, such as real-time PCR or western blotting, should be used to confirm the outcomes of DNA microarray analysis.
It is significant to note that depending on the setup of the experiment and the platform employed, the particular procedure for DNA microarray analysis may change. To achieve accurate and trustworthy results, researchers should carefully adhere to any published protocols or directions from the manufacturer.
Types of DNA microarray
There are two main types of DNA microarray: oligonucleotide microarrays and cDNA microarrays.
Oligonucleotide microarrays
Short, single-stranded DNA pieces, usually 25–70 nucleotides long, make up oligonucleotide microarrays. Every oligonucleotide probe has been created to complement a particular gene or portion of a gene. It is possible to create oligonucleotide microarrays utilizing inkjet printing or photolithography. Due to the exact control over probe length and composition, they are more specific and reproducible than cDNA microarrays.
cDNA microarrays
Cloned cDNA fragments, which are generally 500–2000 nucleotides long, make up cDNA microarrays. Reverse transcription is used to create the cDNA fragments from mRNA, and they are subsequently adhered to a stable surface, such a glass slide or membrane. The quantity of mRNA for each gene is detectable via hybridization, and each cDNA probe represents a different gene. Due to the varied lengths and make-up of the cDNA fragments, cDNA microarrays are less accurate and repeatable than oligonucleotide microarrays.
The choice of microarray platform relies on the particular research issue and experimental setup. Both forms of microarrays offer benefits and drawbacks. Although cDNA microarrays may be better suited for identifying changes in gene expression levels over time or in response to various treatments, oligonucleotide microarrays are often favored due to their superior specificity and repeatability.
Differences between cDNA microarrays and Oligonucleotide microarrays
| Feature | cDNA Microarray | Oligonucleotide Microarray |
| Probe composition | Cloned cDNA fragments | Short, synthetic single-stranded DNA fragments |
| Probe length | Typically longer (500-2000 nucleotides) | Typically shorter (25-70 nucleotides) |
| Probe specificity | Less specific (may contain sequences from multiple genes) | Highly specific to a particular gene or region |
| Reproducibility | Less reproducible | Higher reproducibility due to precise control over probe composition |
| Signal intensity | Generally higher due to longer probe length and hybridization | Generally lower due to shorter probe length and weaker hybridization |
| Sensitivity | More sensitive due to longer probe length and higher signal-to-noise ratio | Less sensitive |
| Specificity | Less specific due to potential cross-hybridization | Higher specificity due to designed probes |
| Cost | Generally, less expensive | Generally, more expensive |
| Design flexibility | Limited due to fixed probes on the array | More flexible due to the ability to custom design probes |
| Data analysis | Analysis methods can be complex and require careful normalization | Easier to analyze due to highly specific probes and more consistent data |
| Application | Useful for detecting changes in gene expression levels over time or in response to different treatments | Useful for high-throughput screening and for studying known or annotated genes |
Applications of DNA Microarray Technology
DNA microarray technology has a wide range of applications, including:
- Gene expression analysis: DNA microarrays are often used to compare the levels of gene expression in various samples, such as healthy and sick tissues, in order to pinpoint the genes that are up- or down-regulated under particular circumstances.
- Genotyping: Single nucleotide polymorphisms (SNPs), which are linked to illnesses or characteristics, can be identified through DNA microarrays.
- Drug discovery: DNA microarrays can be utilized to identify compounds that target certain genes or pathways and to evaluate possible therapeutic candidates for their influence on gene expression.
- Microbial analysis: Based on their genetic profiles, DNA microarrays may be used to identify and categorize microorganisms.
- Forensic analysis: DNA microarrays can be used in paternity tests or criminal investigations to identify people based on their genetic characteristics.
Impact of DNA Microarray Technology
DNA microarray technology has had a significant impact on various fields of science, including:
- Medical research: DNA microarrays have transformed the study of diseases by enabling scientists to pinpoint the genes and biochemical pathways that contribute to a wide range of illnesses, including cancer, cardiovascular disease, and neurological disorders.
- Personalized medicine: With the use of DNA microarrays, clinicians may now customize medicines for specific patients in light of their genetic profiles, opening up new possibilities for customized medicine.
- Agricultural research: DNA microarrays have been used to increase agricultural yields and create new strains of crops that include desired features like insect resistance or a tolerance for environmental challenges.
- Environmental monitoring: DNA microarrays have been used to track the diversity and activity of microbes in a variety of environmental samples, including soil and water, revealing information on how human activities affect the environment.
The study of gene expression has been transformed by DNA microarray technology, which has also created new opportunities for drug development, environmental monitoring, and customized treatment. It has an unquestionable influence on many scientific domains, and it has a huge potential for development. We may anticipate even more fascinating uses and discoveries in the future as technology develops.