History
The trend set by Southern blotting (in 1975) to detect specific DNA brought new ideas in the field of modern molecular biology.
The method got modified in 1977, to develop something very similar to the southern blot when James Alwin, David Kemp and George Stark at Stanford University repeated the design of the southern blot. The major difference was the use of RNA sample to detect a specific RNA molecule within that sample.
This was performed to transfercellular RNA to chemically activated cellulose paper and given the name northern blot for detection of RNA. However, the technique still made use of a radio-labelled DNA probe.
Definition of Northern Blotting
The technique that is used in molecular biology research to study gene expression by detection of RNA or isolated mRNA in a sample is called northern blotting (RNA blotting). It is a classical method for analysis of the size and steady state level of a specific RNA in a complex sample.
It is relatively simple to perform, inexpensive and not obstructed by artefacts. However, there are many technical difficulties that may be encountered in northern blotting because of the process of gel fractionation of the RNA and probe preparation to variation in the quality of the blotting membrane utilized.
Principle of Northern Blotting
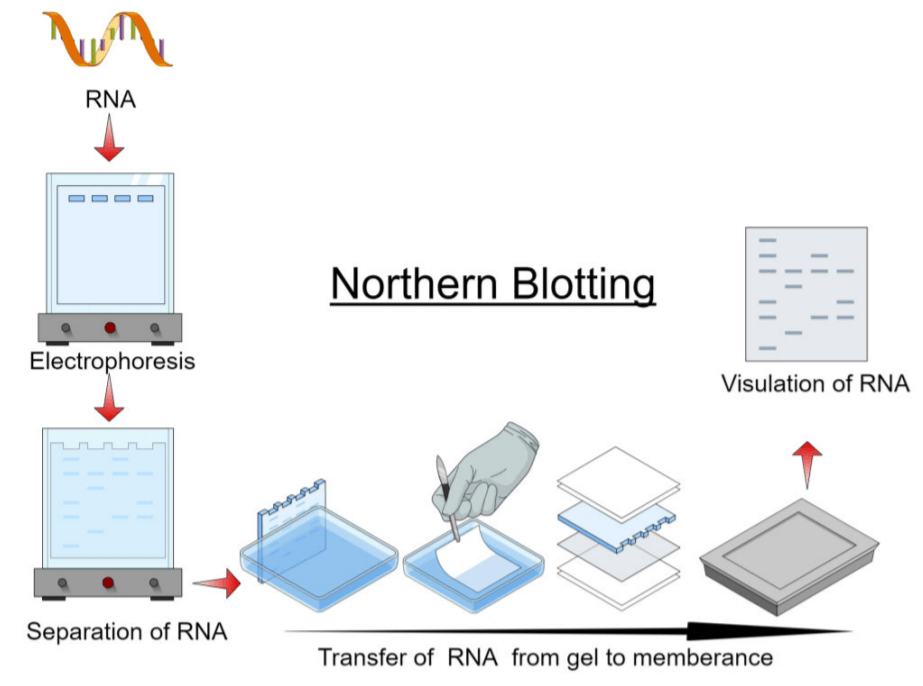
The fundamental principle of northern blotting is to separate RNA based on their size using gel electrophoresis and identified on a cellular membrane by means of hybridization probe with a base sequence corresponding to all or a part of the chain of the target RNA.
- Initially, we extract RNA from a tissue through chaotropic agents such as guanidinium isothiocynate to cause disrupting in cells and denaturing the proteins as well as dissolving the RNA. Some cases require the isolation of mRNA from total RNA using a poly-A+ selection procedure.
- The derived RNA then undergoes agarose-gel electrophoresis where it gets separated which is then proceeded by blotting onto a nylon membrane.
- The RNA on the membrane must be immobilized after blotting through baking or exposure under UV light avoiding nucleic acid to wash away later.
- Finally, hybridization probe is made ready and the probe is hybridized with the membrane.
- A post hybridization wash is required so that the probe is bounded to target mRNA or not is ensured.
- The signals are then detected and visualized using x-films and other methods.
Northern Blotting Procedure
1. RNA Isolation
- Multiple ways of isolating RNA but all have some common attributes such as cellular lysis and membrane disruption, inhibiting of ribonuclease activity, deprotenization and recovery of intact RNA.
- Some of the common methods of RNA extraction are:
RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction.
RNA isolation by column based technology.
- Assessment of quantity and quality of RNA through spectrophotometry.
2. Separation of RNA using gel Electrophoresis
- RNA have secondary structure formed by intramolecular base pairing that prevents RNA from separation according to their size.
- Denaturing agent (formaldehyde or glyoxal/DMSO) disrupts the secondary structure.
- Separation of RNA is better in glyoxal/DMSO system as sharper band for specific RNA is detected by hybridization.
- Size of RNA fragments are compared by migration distance with those of molecular weight markers.
- Gels are stained with ethidium bromide after electrophoresis to detect molecular markers and rRNAs.
3. Transfer of RNA to a membrane
- Nitrocellulose and nylons are commonly used membranes. (Nylon membrane are more durable)
- RNA are transferred via capillary or vacuum transfer. Vacuum transfer are more efficient but require special transfer apparatus.
- Electrophoretic transfer method is available only for nylon membrane due to low concentration of salts required to bind the RNA.
- Transferred RNA are then immobilized to membrane by baking at 80 °C or through UV crosslinking in case of nylon membrane.
4. Hybridization and Washing
- Hybridization is performed using radio or fluorescently labelled probe to identify specific RNA immobilization.
- Pre-hybridization blocks single stranded probe from binding on non-specific sites on the membrane.
- Hybridization solution should contain 50% formamide to ensure hybridization at lower temperature and minimize RNA degradation.
- The membrane is washed in the buffer containing lower concentration of salt to remove excess probes.
5. Visualization
- Detection of specific transcript through autoradiography.
- Membranes are place over X-ray film.
- The X-ray film darkens where fragments are corresponding to the radioactive probes.
Applications of Northern Blotting
- Standard for studying and inspecting gene manifestationdesign between tissues, organs, developmental stages, pathogen infection, and over the course of treatment.
- Diagnosis of several diseases (Crohn’s diseases) including viral infection.
- Exhibit overexpression of oncogenes and down regulation of tumor suppressor genes in cancerous cells.
- Study of RNA degradation and splicing.
- Detect specific mRNA molecular weights and contents in a sample.
- Identify mRNA produced by the transgene to protect recombinants
Advantage and Disadvantages
Advantages
- Relatively simple, cost effective, reduced artifacts.
- Spotted membranes can be stripped of the probes and are reused for hybridization.
- Detect slightest of gene expression changes due to its sensitivity.
- Due to dilution and housekeeping genes control, the results are highly reliable.
Disadvantages
- Time consuming (Only one gene can be analyzed at a time).
- RNA degradation risks because of RNases contamination in work environment.
- Relatively expensive for large scale analysis as huge amount of RNA and reagents are required.
References
- Kenneth M.Rosen, Edward D. Lamperti, Lydia Villa-Komaroff (1990), Optimizing the Northern Blot Procedure
- Paul Trayhurn (1996), Northern blotting
- Akhira Muto, Ken-ichi Arai (1998), Northern Blotting
- Terry Brown, Karol Mackey, Tingting Du (2004), Analysis of RNA by Northern and Slot Blot Hybridization
- Gurman S Pall, Andrew J Hamilton (2008), Improved northern blot method for enhanced detection of small RNA
- KnudJosefsen, Henrik Nielsen (2011), Northern Blotting Analysis
- D Lovatt, J Eberwine (2013), Northen Blotting
- M.W. Nicholas, Kelly Nelson (2013), North, South, or East? Blotting Techniques
- Khaled Moustafa, Joanna M.Cross (2016), Genetic Approaches to Study Plant Responses to Environmental Stresses
- AlshammariFanar Hamad, Hye-Young Jeong, Jong-Hun Han and Irfan A. Rather (2017), Detection of cytosolic tRNA in mammal by Northern blot analysis