Preserving genomic sequence information in living organisms is incredibly important for the perpetuation of life. It’s known that DNA, the essential unit of inheritance, is a reactive molecule and incredibly vulnerable to chemical modifications by endogenous and exogenous agents. However, cells are equipped with systems: DNA repair, damage tolerance, cell cycle checkpoints, and apoptosis pathways that collectively function to chop back the deleterious consequences of DNA damage. DNA repair mechanisms act to prevent mutation and necrobiosis, thus maintaining genome stability, cellular integrity and preventing the onset of cancer, neurodegeneration, and premature aging.
The different categories of DNA repair system that the cells possess are:
1. Direct repair system
Act directly on damaged nucleotides, converting each back to its original structure. Only some kinds of damaged nucleotides can be repaired directly.
Nicks
- In often cases, nicks result from radiation.
- Nicks with broken phosphodiester bond, without any damage to the 5’phosphate and 3’hydroxyl groups, are often repaired by DNA ligase.
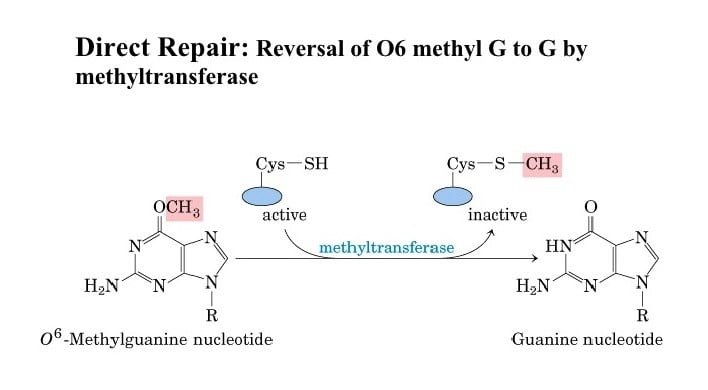
Alkylation damage
- Some forms of alkylation damage can be directly reversed by enzymes.
- In mammals, MGMT or AGT repairs two varieties of DNA adducts: O6-methylguanine (O6-meG) and O4-methylthymine (O4-meT).
- Removal of O6-meG and O4-meT modifications are achieved via a one-step methyltransferase reaction.
- MGMT accepts the alkyl adduct from the modified oxygen molecule, directly restoring the DNA base and inactivating itself.
2. Excision repair
The human genome sequence contains just one gene coding for a protein involved in direct repair (MGMT gene) but has a minimum of 40 genes for components of the excision repair pathways. These pathways represent two categories:
Base excision repair (BER)
- It involves the removal of a damaged nucleotide base, excision of a brief piece of the polynucleotide round the AP site thus created, and resynthesis with a DNA polymerase.
- The process is initiated by an enzyme, DNA glycosylase that recognizes the damaged nucleotide and cleaves the ß-N-glycosidic bond between a damaged base creating an AP site.
- AP endonuclease (APE1 in humans) then cuts the phosphodiester bond on the 5′ side of the gap. Some remove the sugar from the AP site while others lack this ability and work with a separate phosphodiesterase to get rid of it.
- Bifunctional DNA glycosylase can convert a base lesion into a single-strand break without the need for an AP endonuclease.
- The single nucleotide gap is then filled in by a DNA polymerase ß in humans, and the ultimate phosphodiester bond is put in place by a DNA ligase I.
Nucleotide excision repair (NER)
- A long stretch of a polynucleotide (24–29 nucleotides in humans) is removed in this process.
- The bulky lesions are directly recognized by XPC-RAD23B, which binds the non-damaged strand opposite the lesion.
- TFIIH interacts with XPC-RAD23B and opens the DNA with its XPB subunit allowing XPD to trace along with DNA until it stops at the damage and verifies the lesion.
- Stalling of XPD at the lesion allows for the formation of the pre-incision complex by recruitment of XPA, RPA, and XPG. The endonuclease XPG doesn’t make an incision at this time.
- Recruitment of ERCC1-XPF by interaction with XPA to the complex ends up in incision at 5′ of the lesion.
- Initiation of repair synthesis by DNA Polymerase δ/κ/ε and associated factors, followed by 3′ incision by XPG, and sealing of the nick by DNA ligase completes the process.

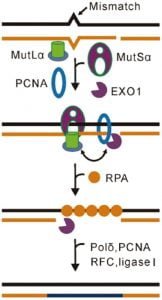
3. Mismatch repair
- A DNA mismatch generates due to the incorporation of an incorrect base during the DNA replication process.
- MutS𝛼 recognizes base-base mismatches, and MutL𝛼 nicks the 3’ or 5’ side of the mismatched base.
- The nicked DNA segment is then excised by the EXO1 exonuclease, in cooperation with the single-stranded DNA-binding protein RPA.
- DNA polymerase 𝛿 and DNA ligase resynthesize the DNA strand.
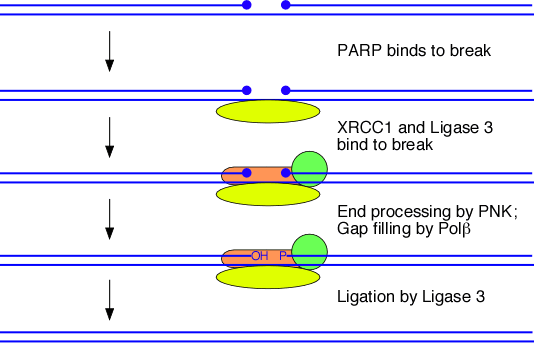
Single-stranded break (SSB) repair
- SSBs are generated due to oxidative damages. They, however, do not give a critical problem to the cell as the double helix retains its overall intactness.
- The single-strand with a break gets coated with PARP1 proteins.
- It protects the strand from breaking and prevents it from participating in unwanted recombination.
- The break is then filled in by the enzymes involved in the excision repair pathways.
4. Double-stranded break (DSB) repair
Various chemical and physical DNA damaging agents induce toxic DSBs which are involved in different human disorders and cancers.
Non-homologous end-joining
- The Ku complex is the first to recognize break and bind to the ends.
- The complex is a heterodimer, includes two copies of the Ku protein (Ku70, Ku80), one copy attaching to each broken DNA end.
- Individual Ku proteins have an affinity for each other and bring the cut ends of the DNA molecule closer.
- Ku binds to the DNA in association with the DNA-PKCS, activates XRCC4, which interacts with the mammalian DNA ligase IV and DNA polymerase ß, to assist in double-strand break repair.
Homologous recombination
- During homologous recombination, the repair of DSB is initiated with the resection of the DNA ends by the combined action of the MRN complex and CtIP generating single-stranded DNA.
- DNA binding proteins RPA, BRCA1, BRCA2, and RAD51 bind to the single-stranded ends and invade the homologous template.
- In subsequent steps, a Holliday junction is generated to complete the DNA synthesis and restore genetic information that was disrupted.

Abbreviations
- AGT: O6-alkylguanine-DNA alkyltransferase
- APE1: Apurinic/apyrimidinic Endonuclease 1
- BRCA: Breast cancer
- CtBP: C-terminal-binding protein
- CtIP: CtBP-interacting protein
- DNA: Deoxyribonucleic Acid
- DNA-PKCS: DNA-dependent protein kinase, catalytic subunit
- EXO1: Exonuclease 1
- ERCC1: Excision repair 1, endonuclease non-catalytic subunit
- MGMT: O6-methylguanine-DNA methyltransferase
- MRN: Mre11/Rad50/Nbs1
- PARP: Poly (ADP-ribose) polymerase
- RPA: Replication protein A
- TFIIH: Transcription factor II Human
- XPB: Xeroderma pigmentosum, complementation group B
- XRCC: X-ray repair cross-complementing protein
References
- Brown TA. Genomes. 2nd edition. Oxford: Wiley-Liss; 2002. Chapter 14, Mutation, Repair and Recombination. Available from: https://www.ncbi.nlm.nih.gov/books/NBK21114/
- Brochier C, Langley B. Chromatin modifications associated with DNA double-strand breaks repair as potential targets for neurological diseases. Neurotherapeutics. 2013;10(4):817-830. doi:10.1007/s13311-013-0210-9
- Fukui K. DNA mismatch repair in eukaryotes and bacteria. J Nucleic Acids. 2010;2010:260512. Published 2010 Jul 27. doi:10.4061/2010/260512
- Schärer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5(10):a012609. Published 2013 Oct 1. doi:10.1101/cshperspect. a012609