What is Pulsed field Gel Electrophoresis (PFGE)?
Pulsed – field gel electrophoresis is a separation technique of large DNA molecules (entire genomic DNA) to produce a DNA fingerprint for bacterial isolates. It is a powerful genotyping technique that the DNA molecules are digested with unique restriction enzymes and are separated under the electric field that changes direction periodically.
Principle of Pulsed field Gel Electrophoresis:
The main principle of gel electrophoresis is to separate the fragments of DNA according to their size under the electric field. In pulsed field gel electrophoresis, large DNA molecules with above 30 to 50 kb length fragment runs on the gel matrix at the same rate and appears as a single large diffuse band.
The various length of the DNA changes with the periodically changing direction of electric field where large DNA molecule moves slowly when the direction of field is changed while small DNA molecule moves faster. This separation of molecule continues over the course of time with the consistent changing of direction.
Why pulse field gel electrophoresis over standard gel electrophoresis technique?
The standard gel electrophoresis only separates small molecules of DNA and is unable to separate DNA molecules larger than 15 to 20kb effectively. In 1984, David C. Schwartz and Charles Cantor developed an electrophoresis technique that can separate the DNA molecules larger than 50kb which became known as pulsed field gel electrophoresis. The development of PFGE provided a huge advantage for molecular biology research and expanded the range of resolution for DNA fragments.
How does PFGE work?
The concept of standard gel agarose electrophoresis and pulsed field gel electrophoresis is similar but equipment required to run PFGE is much more complicated and needs a trained personnel.
Following are the procedure for performing pulsed field gel electrophoresis:
- Bacterial cells are taken from an agar plate and are mixed and loaded with melted agarose.
- Bacterial cells are immobilized into agarose blocks known as plug mold to protect the chromosomal DNA from mechanical damage.
- This is the first step where bacterial cells are lysed and DNA is released in the agarose plug.
- The bacterial DNA is digested with restriction enzymes to produce less number of larger size DNA fragments.
- The digested large sized DNA fragments are subjected to the pulsed field gel electrophoresis where the electric charges are passed from different directions at regular interval where the DNA fragments are separated on the basis of their size.
- The gel is stained with a SYBR Green I where the DNA are visualized under the ultraviolet (UV) light.
- The bands of the fragment of DNA molecules of different organisms are analyzed and compared to standards manually.
- Computer software like BioNumerics is also used.
- A digital camera is also used to take a photograph of the gel.
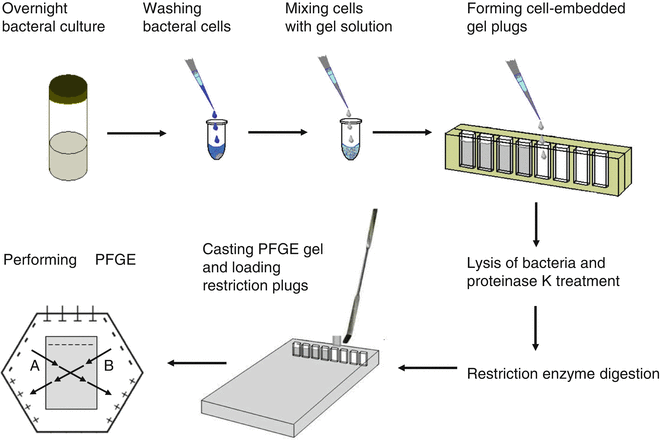
(Source:https://link.springer.com/protocol/10.1007/978-1-4939-2599-5_14)
Components to be enhanced while performing PFGE for the best results
- Voltage: Usually 6 V/cm is used as a standard for a molecule with a few hundred kb size, but if the sample size is larger in mega bases, lower voltage is suggested to use and for small sample size, the voltage used is high.
- Pulse angle: Most protocols usually use 120° angle which can be adjusted in terms of the size of the DNA fragments. To increase the resolution of large fragments, small angels should be used similarly for small fragment, angle should be larger.
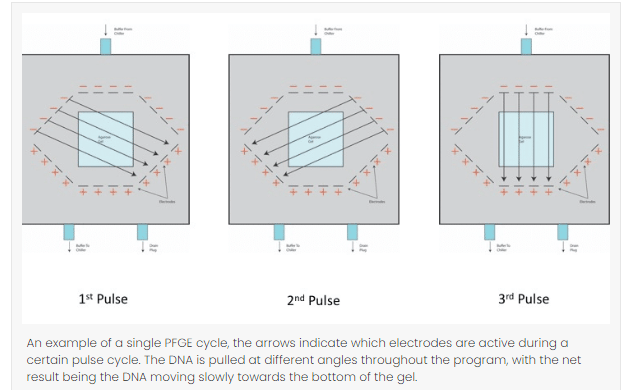
- Switch time: Switch time refers the duration of time the electric current uses to pull in one direction. Short switch time should be used for small sizes and a long switch time for long sized fragments.
- Temperature: To run a PFGE cycle in a gel matrix, it usually takes overnight or a couple of days to complete. Therefore, buffer temperature must be maintained throughout the procedure, a lower temperature takes a longer run time but gives a greater resolution and a higher temperature takes a shorter run time but yields low resolution.
Advantages of Pulsed field gel electrophoresis:
- PFGE hold the strength of separating large DNA molecules to over 10 Mb pairs.
- PFGE is used in the sub typing of many pathogenic bacteria and are mainly used in epidemiological field.
- PFGE follows the similar basic format as a universal generic method for bacterial sub typing and only requires a choice of restriction enzymes and condition of electrophoresis for each sample species.
- The restriction patterns generated by PFGE are stable and can be reproduced.
Disadvantages of pulse field gel electrophoresis:
- PFGE method consumes more time which might take overnight or a couple of days.
- It cannot separate the fragments in every part of the gel at the same time.
- One change in restriction site might form more than one band.
- Not all strains can be sub typed by PFGE.
- PFGE does not differentiate isolates to the same degree as whole genome sequencing (WGE).
Application of pulse field gel electrophoresis:
- PFGE is considered as a gold standard technique in the area of epidemiological studies of pathogenic organisms.
- In genome size estimation, PFGE is considered as an efficient method.
- PFGE is also used for genotyping or genetic fingerprinting.
- Yeast Artificial Chromosome (YAC) libraries are constructed by PFGE.
- PFGE has also been used in the analysis of large DNA molecules from fungi and parasitic protozoa.
- PFGE is also used in the study of radiation-induced DNA damage and repair.
References:
- https://bitesizebio.com/29971/pulsed-field-gel-electrophoresis/
- https://www.cdc.gov/pulsenet/pathogens/pfge.html
- https://microbenotes.com/pulsed-field-gel-electrophoresis-pfge/
- https://www.slideshare.net/jitenderanduat/pulsed-field-gel-electrophoresis-pfge