The real-time polymerase chain reaction is also known as quantitative PCR or qPCR or quantitative reverse – transcriptase PCR which is used in the measurement of a small amount of gene expression which is monitored by PCR reaction in real-time. We will learn about Real-time PCR in detail here.
- RT – PCR can assess multiple PCR simultaneously on a camera or detector detects the amplicon but does not rely on electrophoresis or densitometry.
- It can either be quantitative or semi-quantitative.
- Types of RT-PCR are used on the basis of a study that needs to be done. For instance, basic qPCR is used for gene detection only similarly, reverse transcriptase PCR is used to study the expression of genes.
- To decide which method should be used for the experiment, we should have knowledge about the sample and the result that we desire.

- DNA binding dye is an easy and rapid method that can carry out even by an inexperienced person. The dye binds to the double-stranded DNA and its fluorescence emits by 100 to 1000 fold in an increasing manner. Then, it is taken as the baseline for detection.
- Probe-based method can use multiple probes for multiple template amplification. It is of two types: Linear probe (Taqman probe) and molecular beacons.
Basic principle of RT-PCR
- Real-time PCR process is generally carried out for the detection of fluorescence emitted by the excited molecules which increase as the reaction proceeds.
- It is carried out in a thermal cycler which has a capacity to illuminate each molecule with at least one wavelength of light.
- The fluorescent molecule appears due to the binding of the dye to the ds-DNA or sequence-specific probes. The dye used is SYBR Green and probes are molecular beacons or Taqman probe.
- For the visualization of amplified DNA fragments, non-specific fluorescent DNA dyes, and fluorescently labeled oligonucleotide probes are included for the pathogen detection.
Steps and procedures of RT-PCR
RT – PCR is completed in two major steps:
- The first step includes the conversion of RNA to complementary DNA (cDNA) which can be achieved by amplifying and monitoring the sample.
- The second step is the use of fluorescent reporters like the SYBR Green and TaqMan probe which are the two commonly used reporters that help in the detection of a specific gene from the sample.
As like the conventional PCR, amplification in RT-PCR is done in three steps:
Denaturation:
A high temperature of about 94°C melts and denatures the double-stranded DNA into two single strands. Denaturation time depends upon the GC content of DNA.
2. Annealing:
The sequence-specific primer and the fluorescent dye or probe binds to the single-stranded DNA sequence at 55 to 66°C.
3. Extension:
Extension occurs at 70 to 72°C where the primer extension is highest at this temperature. But in real-time PCR, an amplicon is small and is often combined with the annealing step at 60°C.
The next step is the detection of specific genes from the sample using SYBR Green and Taqman probe.
- SYBR Green is a fluorescent dye that binds between the DNA bases which determines the amount of DNA that has been amplified.
- Each TaqMan probe binds in between the forward and reverses primer while the quencher and reporter bind to the probe and the quencher gets separated from the reporter dye allowing the fluorescence to be detected.
- The detection of a specific gene is based on fluorescence technology.
- The placed sample is subjected to a tungsten or halogen source where the signal is amplified.
- These signals are detected by a detector and are displayed on a computer screen.
How SYBR green works?
- SYBR green is a green fluorescent cyanine dye that binds to the double-stranded DNA molecule and emits a prominent fluorescent signal.
- On the denaturation of the DNA molecule, the SYBR green molecule gets released and reduces the fluorescent drastically.
- During polymerization, primer anneal and PCR product is generated then SYBR green binds to the DNA bases which results in an increase of fluorescence.
- SYBR green dye is preferred over other dye because of its high signal intensity but lacks specificity when compared to the Taqman probe.
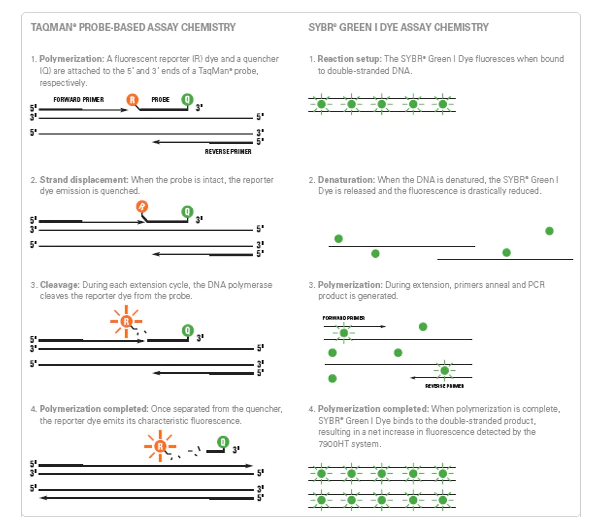
How Taqman probe works?
- Taqman probe is a fluorescently labeled DNA oligonucleotide whose 5’ end is labeled with a fluorescent reporter molecule whereas 3’ end is a quencher molecule.
- When the probe becomes intact, the reporter dye emission gets quenched and DNA polymerase cleaves the reporter dye during each extension cycle.
- The reporter dye emits its fluorescence signal once gets separated from the quencher.
- As the amplification cycle increases, the signal fluorescence molecule also increases.
Application of Real Time-PCR
- RT-PCR is used in biomedical science research and nanotoxicology studies for semiquantitative analysis of target genes.
- RT-PCR is also applicable in the field of clinical microbiology laboratories for the diagnosis of human microbial infections.
- It is also useful in the detection of cancer cells and analyzes its expression level.
- It is also applied in food industries for genetically modified organism (GMO) food testing and water quality.
- In the agricultural field, it helps in the detection of phytopathogens.
- In the pharmaceutical industry, it is useful in drug research and helps in detecting the anti-inflammatory effect of peptides.
- Real time PCR also helps in forensic medicines.
Learn more:
Refrences:
- https://en.wikipedia.org/wiki/Real-time_polymerase_chain_reaction#:~:text=Real%2Dtime%20PCR%20permits%20the,used%20is%20usually%20SYBR%20Green.
- https://www.ebi.ac.uk/training-beta/online/courses/functional-genomics-ii-common-technologies-and-data-analysis-methods/real-time-pcr/
- http://www.premierbiosoft.com/tech_notes/real_time_PCR.html
- https://www.frontiersin.org/articles/10.3389/fmicb.2017.00108/full
- Maddocks, S., & Jenkins, R. (2017). Quantitative PCR. Understanding PCR, 45–52.
- https://geneticeducation.co.in/real-time-pcr-principle-procedure-advantages-limitations-and-applications/
- https://www.thermofisher.com/np/en/home/life-science/pcr/real-time-pcr/real-time-pcr-learning-center/real-time-pcr-basics/taqman-vs-sybr-chemistry-real-time-pcr.html
- https://microbenotes.com/real-time-pcr-principle-process-markers-advantages-applications/
- http://www.aun.edu.eg/molecular_biology/Procedure%20of%20PCR%20level%202%20(5.7-4-2016)/RT-PCR%20application%20(1).pdf