What are single nucleotide polymorphisms (SNPs)?
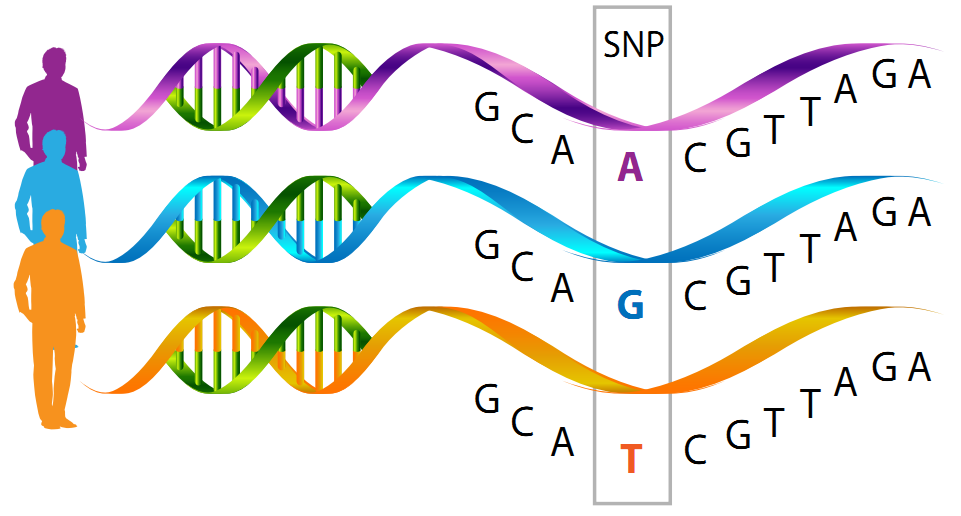
A single nucleotide polymorphism (SNPs) (Pronounced: snip) is a DNA sequence variation that arises when a single nucleotide (adenine, thymine, cytosine, or guanine) in the genome sequence is altered and the specific modification is present in at least 1% of the population.
When a single nucleotide, such as an A, substitutes one of the other three nucleotide letters – C, G, or T – this is referred to as SNP variation.
According to the traditional definition of polymorphism, the frequency of the variation must be at least 1% to define the nucleotide change as a polymorphism. Rare variants are nucleotide alterations that occur in fewer than 1% of the population.
Not all SNP have a major impact on human health or function. Numerous SNPs are found in non-coding areas of the genome or in locations where gene expression or protein function are not directly impacted. SNP that are found in the genes’ regulatory or coding areas, however, might have functional ramifications that affect phenotypic features, illness risk, or treatment responsiveness.
Origin of SNPs
Single Nucleotide Polymorphisms (SNPs) can arise through various mechanisms, both natural and induced. Here are the primary origins of SNP:
DNA Replication Errors:
- Errors in nucleotide sequence copying can occur.
- DNA polymerase, the enzyme responsible for replicating DNA, can at times insert the erroneous nucleotide or skip a nucleotide, leading to a substitution, insertion, or deletion of a single nucleotide.
Spontaneous Mutations:
- SNPs can develop on their own spontaneously without any external trigger.
- Mutations can occur as a result of chemical changes inside the DNA molecule or mistakes in DNA repair mechanisms.
- Exposure to environmental mutagens or the intrinsic instability of particular genomic areas can all contribute to the formation of spontaneous SNPs.
Mutagen Exposure:
- Exposure to certain environmental factors, known as mutagens, can increase the likelihood of SNP formation.
- Mutagens include radiation (e.g., ultraviolet light, ionizing radiation), certain chemicals (e.g., carcinogens, toxins), and some viruses.
Inherited SNPs:
- SNPs can be inherited from parents.
- When germ cells (sperm and egg cells) go through meiosis, they can pick up SNP from their parents’ DNA.
- Inherited SNPs can be handed down across generations, contributing to genetic diversity within a community.
SNP have several sources, including extrinsic variables like exposure to mutagens as well as intrinsic causes like spontaneous mutations and mistakes in DNA replication. Understanding the natural processes and variables that contribute to genetic diversity and variation within populations is made possible by the study of SNP origins.
Significance of SNPs
Single Nucleotide Polymorphisms (SNPs) are important because they can act as genetic markers for several facets of human life. Here are some crucial details emphasizing the importance of SNPs:
- Genetic Variation: SNPs are the most prevalent type of genetic variation, occurring in the human genome around once every 300 nucleotides. They play a significant part in evolution and adaptation, adding to the variety found in both individuals and communities.
- Disease Susceptibility: SNP may be linked to a higher or lower chance of contracting particular diseases. Numerous SNPs have been shown to be associated with a wide range of illnesses, including Alzheimer’s disease, diabetes, cancer, and autoimmune disorders, according to genome-wide association studies (GWAS). Early identification, risk assessment, and customized disease preventive programs can all benefit from an understanding of these relationships.
- Pharmacogenomics: SNPs affect how a person reacts to drugs. Medication effectiveness, medication metabolism, and the likelihood of adverse drug responses can all be impacted by certain SNPs. SNPs are used in pharmacogenomic research to customize medication regimens based on a patient’s genetic profile, improving treatment success and reducing adverse effects.
- Population Genetics: In population genetics research, SNPs are important. SNP comparisons allow researchers to examine ancestry, migration, and genetic variation trends in various populations. Understanding human evolution, migratory trends, and the genetic variety existing in various communities throughout the world is made easier by this knowledge.
- Forensic Identification: SNP may be used in forensic research to identify human beings. Forensic scientists may develop genetic profiles that are distinctive to each person by studying certain SNPs, which can help with criminal investigations and paternity determination.
- Research into evolution: SNPs offer insight on how species have evolved. Researchers may reconstruct evolutionary links and understand the genetic underpinnings of species diversification by analyzing SNPs in a variety of animals to better understand the genetic alterations that have taken place through time.
- Targeted medications: Precision medicine can employ SNP to create tailored medicines. Researchers can create medications that particularly target such genetic variants by finding SNPs linked to certain diseases, resulting in more effective therapies with fewer adverse effects.
- Functional genomics: By changing gene expression, protein structure, and function, SNPs can have functional repercussions. They may change how genes are controlled, resulting in changes in phenotypes and susceptibility to illness. Understanding how SNPs affect function sheds light on the biological processes and pathways involved in the emergence of illness.
In conclusion, SNPs play a crucial role in genetic and genomic research. They aid in forensic identification, enable tailored treatment techniques, shed light on evolutionary processes, and explain the genetic underpinnings of disorders. SNP research provides enormous promise for expanding scientific understanding and improving healthcare while also deepening our understanding of human biology.
Detection of SNPs
Single Nucleotide Polymorphisms (SNPs) are genetic variants that may be found and characterized by researchers using a variety of methods and procedures. The following are some popular techniques for SNP detection:
- Polymerase Chain Reaction (PCR): PCR is a popular method for finding SNP. The SNP region of interest is flanked by specific primers, and genomic DNA is used as a template for PCR amplification. By examining the PCR results using techniques like gel electrophoresis, DNA sequencing, or allele-specific PCR, it is possible to identify whether a certain nucleotide is present at the SNP site or not.
- DNA sequencing: SNP detection frequently makes use of Sanger sequencing and next-generation sequencing (NGS) methods. By comparing the sequence acquired with a reference sequence, SNPs can be identified by Sanger sequencing, which amplifies DNA fragments before sequencing them. Comprehensive SNP identification across the genome is made possible by NGS techniques, such as whole-genome sequencing or targeted sequencing, which can quickly evaluate a huge number of DNA fragments.
- Allele-Specific Oligonucleotide Hybridization (ASO): This hybridization-based method makes use of brief DNA probes that are exclusive to each SNP allele. These probes are designed to exclusively attach to an allele’s complementary sequence. It is possible to establish if each SNP allele is present or absent by examining the probes’ binding to the DNA sample.
- Restriction Fragment Length Polymorphism (RFLP) Analysis: Using restriction enzymes that can detect certain sequences around the SNP, DNA is digested in RFLP analysis. Individuals with different SNP alleles will have variable DNA fragment lengths if an SNP modifies the restriction enzyme recognition site. SNPs can be found by examining the size variation of the resultant DNA fragments using gel electrophoresis.
- Techniques based on Hybridization: Methods that rely on hybridization, such microarray-based genotyping or bead-based assays, use DNA probes that have been fixed on a stable substrate. These probes are intended to collect SNP targets alone. The probes and the labeled DNA sample are hybridized. SNPs can be located by examining the pattern of probe-target binding.
- Mass Spectrometry: Mass spectrometry-based techniques for SNP genotyping include pyrosequencing and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). To find SNP differences, these approaches examine the quantity or organization of nucleotides in a DNA sample.
- High-Resolution Melting (HRM) Analysis: HRM analysis is a PCR-based technique that identifies SNPs by comparing how differently DNA fragments melt. As DNA strands denature, the fluorescence changes, and SNPs may be identified by their distinctive melting curves.
Our understanding of genetic differences and their consequences for numerous biological processes, disease susceptibility, and customized therapy has considerably improved because to these SNP detection approaches.
Learn more