Chromatin Immunoprecipitation (ChIP) is a technique used to study the interactions between proteins and DNA in a specific region of the genome. It is commonly used to investigate how proteins, such as transcription factors or histones, bind to DNA and regulate gene expression.
Chromatin Immunoprecipitation (ChIP) Protocol
The Chromatin Immunoprecipitation (ChIP) technique involves several steps to study protein-DNA interactions in a specific region of the genome. Here are the general steps involved in ChIP:
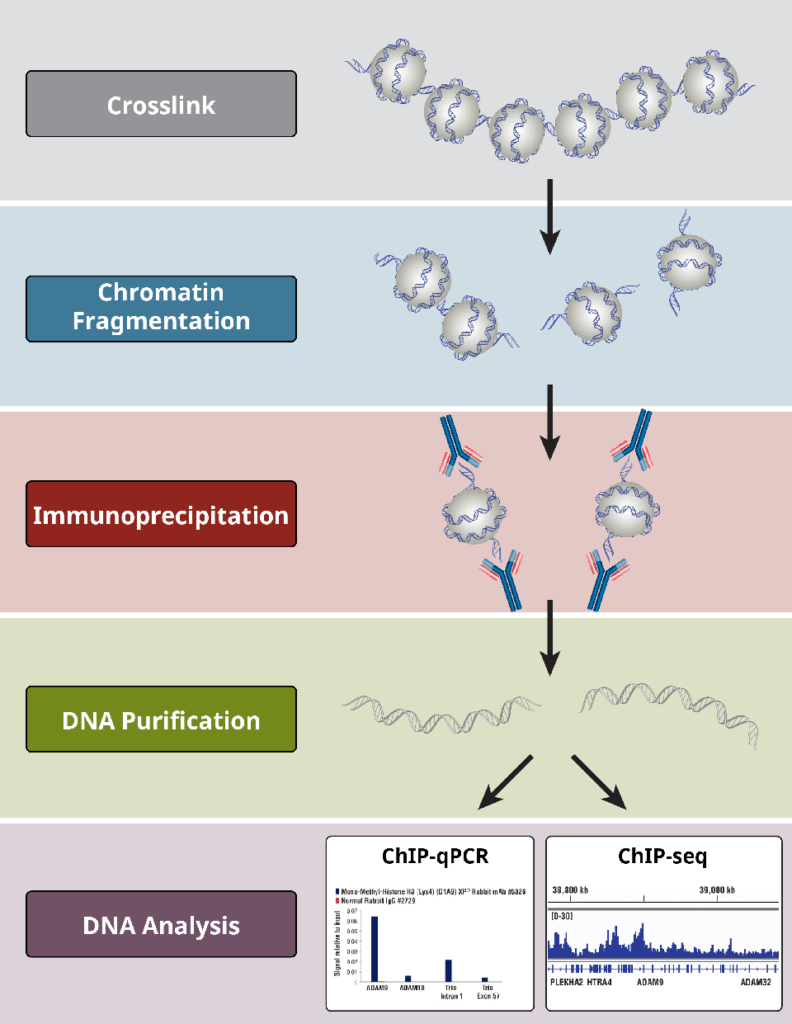
1. Cross-linking
To retain the protein-DNA connections, cells or tissues are treated with a cross-linking agent (often formaldehyde). This chemical cross-linking facilitates in the stabilization of complexes generated between proteins and DNA.
- Collect cells or tissues and wash them in ice-cold phosphate-buffered saline (PBS).
- Add formaldehyde (1% final concentration) to cross-link the proteins and DNA. Incubate for 10 minutes at room temperature with moderate agitation.
- Stop the cross-linking process by adding glycine (0.125 M final concentration). Incubate for 5 minutes at room temperature with moderate agitation.
- Wash the cells or tissues twice with ice-cold PBS and collect them by centrifugation.
2. Chromatin Isolation
The cross-linked cells or tissues are lysed to free the chromatin (DNA-protein complexes). To enable the next stages, the chromatin is sheared or split into smaller fragments. This can be accomplished by sonication (the use of sound waves) or enzymatic digestion.
- Lyse the cells or tissues in a lysis solution containing protease inhibitors. Incubate on ice for 10 minutes.
- Sonicate the lysate or utilize enzymatic digestion to break the chromatin into smaller bits. Confirm that the chromatin has been sheared to a suitable size range (200-1000 base pairs).
- Centrifuge the lysate in order to remove cellular debris, then transfer the chromatin-containing supernatant to a separate tube.
3. Immunoprecipitation
A protein-specific antibody is added to the chromatin mixtures. The antibody binds to the target protein, producing an antibody-protein-DNA complex. This process separates the particular protein-DNA complexes from the rest of the chromatin.
- Pre-clear the chromatin in the supernatant by adding protein A/G beads or magnetic beads. Incubate at 4°C for 1 hour with moderate agitation.
- Centrifuge the mixture to remove the beads, and then transfer the cleaned chromatin to a fresh tube.
- Incubate the chromatin with an antibody specific to the protein of interest overnight at 4°C with moderate agitation.
4. Precipitation
Protein A/G beads or protein A/G-coated magnetic beads are introduced to the chromatin-antibody complex. These beads attach to the antibody-protein-DNA complex and have a high affinity for antibodies. Centrifugation or magnetic separation are used to collect the beads and the associated complexes.
- Incubate the chromatin-antibody mixture with protein A/G beads or magnetic beads for 2-4 hours at 4°C with moderate agitation. The antibody-protein-DNA complexes will attach to the beads.
- Centrifuge or magnetically separate the bead-protein-DNA complexes.
5. Washing
The complexes of beads, proteins, and DNA are washed to eliminate any non-specifically bound proteins or DNA fragments. This procedure helps in the reduction of background noise and the specificity of the isolated complexes.
- To eliminate non-specifically bound proteins and DNA fragments, the bead-protein-DNA complexes were washed progressively with low-salt, high-salt, LiCl, and TE buffers.
- Perform each wash step for the stated time and centrifuge or magnetically separate the beads between washes.
6. Elution
Protein-DNA complexes are normally freed from the beads by reversing the cross-links. This is accomplished by heating the complexes overnight at high temperatures.
- Add elution buffer (including SDS and proteinase K) to the beads and incubate at 65°C for several hours or overnight. This procedure breaks the cross-links and allows the DNA to be released.
7. DNA Purification
The DNA fragments obtained from the elution step are purified to remove any contaminants or residual proteins. This purification step ensures that the DNA is in a suitable form for downstream analysis.
- Purify the DNA using phenol-chloroform extraction or a DNA purification kit.
After the ChIP procedure, the purified DNA can be used for various applications, such as quantitative PCR (qPCR) to measure the enrichment of specific DNA regions or next-generation sequencing (ChIP-seq) to identify genome-wide protein-DNA interactions.
ChIP Applications
Chromatin Immunoprecipitation (ChIP) is a versatile technique that has several important uses in molecular biology and genomics research. Here are some common applications of ChIP:
1. Protein-DNA Interaction Analysis
- ChIP enables the investigation of protein-DNA interactions, assisting in the identification of genomic areas where particular proteins bind.
- Used for understanding gene regulation, transcriptional control, and chromatin structure.
2. Transcription Factor Binding
- Used to identify and characterize transcription factor binding sites in the gene.
- Regulatory networks that regulate gene expression and the functions of certain transcription factors in various biological processes by researching transcription factor binding patterns.
3. Histone Modification Analysis
- Used to analyze histone modifications, which are crucial epigenetic markers involved in gene regulation.
- Can map the distribution of various histone modifications across the genome and learn how they affect chromatin structure, gene activation, and repression.
4. Epigenetic Research
- Used in epigenetics to explore heritable changes in gene expression that do not entail changes in the DNA sequence.
- Aids in the identification of DNA methylation patterns, histone modifications, and chromatin accessibility, revealing how these epigenetic markers impact gene expression, development, and illness.
5. Protein-Protein Interaction Analysis
- Used to investigate protein-protein interactions in chromatin.
- Can explore the composition and dynamics of multi-protein complexes engaged in diverse biological processes by co-immunoprecipitating protein complexes linked with certain genomic regions.
6. Genome Mapping
ChIP-seq, a version of ChIP paired with next-generation sequencing, enables the mapping of protein-DNA interactions throughout the whole genome. On a genome-wide scale, it allows for the discovery of binding sites for diverse proteins such as transcription factors, histones, and chromatin modifiers. This method gives a complete perspective of protein-DNA interactions and aids in the annotation of regulatory and functional regions in the genome.
ChIP Limitations
While Chromatin Immunoprecipitation (ChIP) is a useful approach for researching protein-DNA interactions and gene regulation, it does have certain drawbacks. Here are some of the most common ChIP limitations:
- Cross-Linking Specificity: If the cross-linking process in ChIP is not optimized appropriately, it might introduce artifacts. Cross-linking can cause the development of non-specific protein-DNA complexes, which can result in false-positive findings. It is critical to improve the cross-linking conditions in order to reduce non-specific binding while maintaining specificity.
- Antibody Specificity: The specificity and quality of the antibodies used to target the protein of interest are critical in ChIP. Antibodies with limited specificity or cross-reactivity may produce false-positive findings or fail to effectively capture the intended protein-DNA complexes. To provide trustworthy findings, antibodies must be carefully selected and validated.
- DNA Fragmentation Bias: During the ChIP process, chromatin can be sheared or fragmented, which might cause biases. DNA fragment size and distribution may not be totally indicative of the original chromatin structure. Biases in DNA fragmentation can affect immunoprecipitation effectiveness and subsequent protein-DNA interaction studies.
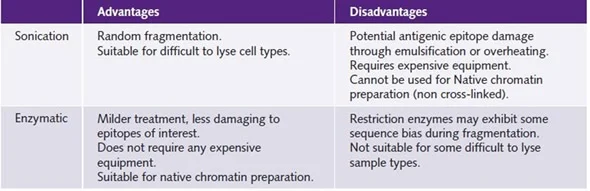
- Epitope Accessibility: ChIP is dependent on the antibody’s ability to reach the protein of interest and the related DNA sections. In other circumstances, proteins may be strongly attached to DNA or placed in physically inaccessible locations, resulting in limited immunoprecipitation effectiveness. This can make detecting and analyzing protein-DNA interactions for specific genomic loci more difficult.
- Background Noise: Non-specific protein or DNA fragment binding can cause background noise in ChIP assay. Non-specific antibody binding, non-specific interactions with chromatin, or non-specific DNA amplification during downstream analysis can all contribute to this noise. To differentiate specific signals from background noise, certain controls and optimization techniques are required.
- Technical Variability: Technical variability in ChIP research can occur as a result of protocol stages such as cell fixation, chromatin fragmentation, immunoprecipitation efficiency, and DNA purification. This heterogeneity can make repeatability difficult and necessitates careful experimental design, standardization, and suitable controls.
To ensure the reliability and interpretation of ChIP data, it is critical to be aware of these limitations and to adopt adequate controls and validation approaches. Combining ChIP with complementary methods and methodologies can also assist to alleviate some of these limitations and give a more thorough knowledge of protein-DNA interactions and gene regulation.
ChIP controls
Chromatin Immunoprecipitation (ChIP) experiments require the inclusion of specific controls to assess the specificity of the immunoprecipitation and to distinguish true signals from background noise. Here are some common controls used in ChIP:
- Negative Control Antibody: A negative control antibody is an antibody that does not recognize the protein of interest or any other relevant protein in the experimental system. This control helps assess non-specific binding of antibodies to chromatin or DNA and provides a baseline for background noise.
- Isotype Control Antibody: An isotype control antibody is an antibody of the same isotype as the primary antibody used in the ChIP experiment but does not recognize any antigen in the sample. It helps to control for non-specific binding of antibodies and assesses background noise due to the immunoglobulin itself.
- No Antibody Control: In this control, no primary antibody is added during the immunoprecipitation step. This control helps evaluate the background signal arising from non-specific interactions between the chromatin and the secondary antibody, beads, or other components of the ChIP procedure.
- Input DNA Control: The input DNA control is a portion of the chromatin sample taken before immunoprecipitation. It represents the total genomic DNA content and serves as a reference for the abundance of the target DNA region in the starting material. The input DNA control is used for normalization purposes during quantitative analysis, such as qPCR or ChIP-seq.
- Positive Control: A positive control is a known target region that is expected to interact with the protein of interest. It can be a well-characterized genomic region known to be bound by the protein or a region known to exhibit specific histone modifications. The positive control helps validate the ChIP protocol and demonstrates that the immunoprecipitation is working effectively.
- Genomic Control Regions: Genomic control regions are regions of the genome that are not expected to be bound by the protein of interest. These regions serve as negative controls to assess the specificity of the immunoprecipitation. The selection of appropriate genomic control regions is important to ensure they represent the general background signal and do not contain any specific binding sites.
Including these controls in ChIP experiments helps assess the specificity of the immunoprecipitation, distinguish true signals from background noise, and ensure the reliability and interpretation of the results. The choice and design of controls may vary depending on the specific experimental setup, target protein, and the goals of the study.
ChIP Troubleshooting
When encountering issues or unexpected results in a Chromatin Immunoprecipitation (ChIP) experiment, troubleshooting can help identify and address the problem. Here are some common troubleshooting steps for ChIP:
Antibody Optimization
- Ensure that the antibody used is specific for the target protein by reviewing literature or conducting validation experiments.
- Consider trying different antibodies or antibody lots to confirm specificity.
- Adjust the antibody concentration and incubation time to optimize the immunoprecipitation efficiency.
Cross-Linking and Chromatin Fragmentation
- Verify that the cross-linking step was performed correctly by assessing the level of cross-linking efficiency. An insufficient cross-linking may result in weak or no signals.
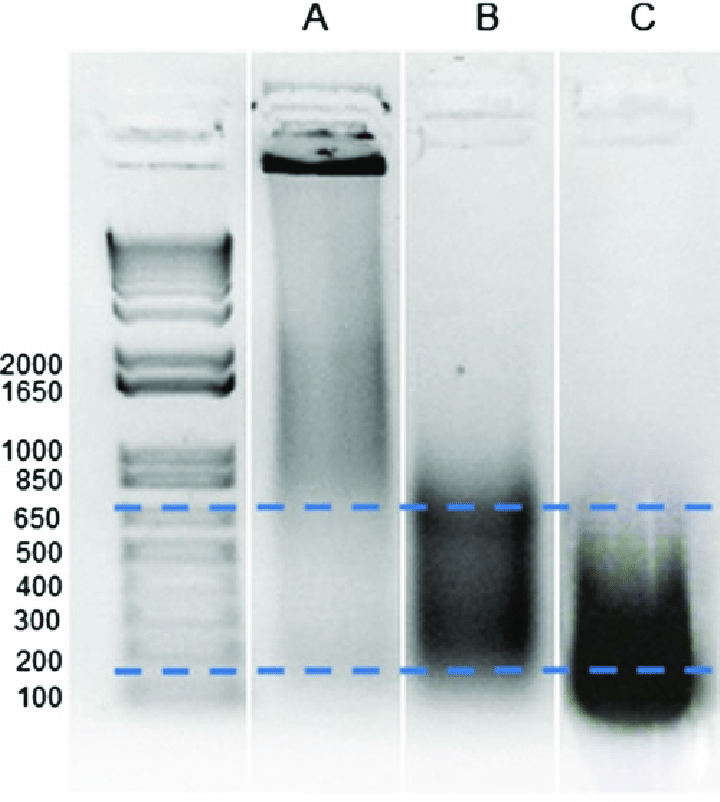
- Optimize chromatin fragmentation to achieve the desired fragment size range (200-1000 base pairs). If fragments are too large or too small, adjust sonication or enzymatic digestion conditions.
- Verify the quality of the sheared chromatin by running a gel or performing qPCR on input DNA samples.
Non-Specific Binding and Background Noise
- Include appropriate negative controls such as a no-antibody control or a negative control antibody to assess non-specific binding and background noise levels.
- Optimize wash conditions to minimize non-specific interactions. Try adjusting the stringency, salt concentration, or detergent concentration during washing steps.
- Evaluate and troubleshoot steps involving blocking reagents, beads, or other components that may contribute to background noise.
DNA Purification and Quantification
- Ensure that DNA purification is performed properly, and contaminants are effectively removed. Impurities in the DNA may interfere with downstream analyses.
- Verify DNA concentration and quality using spectrophotometry or fluorometric assays. Low DNA yield or poor quality may affect subsequent PCR or sequencing steps.
Data Analysis and Interpretation
- Assess the experimental design and statistical analysis methods used. Consider consulting with a bioinformatics expert for data analysis if performing ChIP-seq.
- Validate ChIP results using independent techniques such as qPCR or alternative ChIP methodologies.
- Compare results with existing literature or databases to ensure consistency and interpretation of the findings.