Gene expression is the process by which the information contained in a gene is utilized to control the synthesis of a functional gene product. A complex process, gene expression requires the coordination of several dynamic events and is governed at various levels. These regulatory levels include transcriptional, post-transcriptional, translational, and post-translational levels.
Gene transcriptional regulation is a fundamental part of both tissue-specific gene expression and gene activity in response to stimuli. Transcription factors (TFs) are the primary controllers of gene transcription. A transcription factor (TF) is a protein that can bind to a particular DNA sequence and regulate transcription.
In order to control the rate of gene transcription, transcription factors bind to DNA-regulatory sequences (enhancers and silencers), which are typically found in the region 5’ upstream of target genes. This may lead to alterations in protein synthesis, gene transcription, and consequent cellular function.
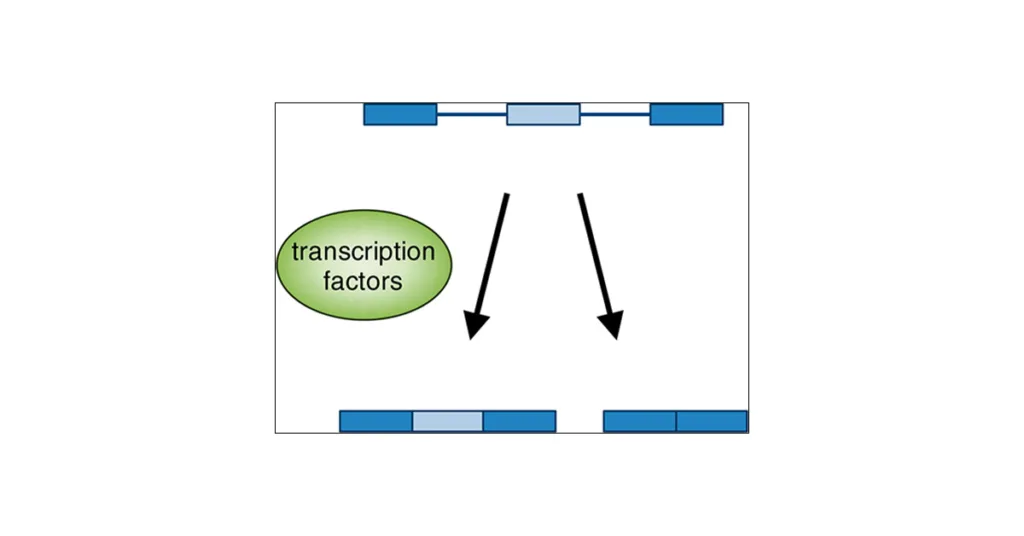
Signal transduction pathways regulate the activation or inhibition of many transcription factors. By creating complexes that can stimulate or repress transcriptional activity, co-factors can also control these processes.
Transcription factors family
There are several families of transcription factors, and members within each family may have similar structural traits. These families include the following:
- helix-turn-helix (e.g., Oct-1),
- helix-loop-helix (e.g., E2A),
- zinc finger (e.g., glucocorticoid receptors, GATA proteins),
- basic protein-leucine zipper [cyclic AMP response element binding factor (CREB), activator protein-1 (AP-1)],
- β-sheet motifs [e.g., nuclear factor-κB (NF-κB)]
Helix-turn-helix
Helix-turn-helix is a DNA-binding protein (DBP). The primary structural motif known as the helix-turn-helix (HTH) can bind to DNA. Each monomer has two α-helices that bind to the main groove of DNA and are connected by a brief strand of amino acids. Many proteins that control the expression of genes have the HTH motif. Archaea, bacteria and eukaryotes utilize HTH in general and basal transcription.

A few examples of proteins with the helix-turn-helix motif are shown below:
- Homeodomain proteins: These proteins are involved in the regulation of gene expression during development. Examples include the Antennapedia protein and the Engrailed protein.
- Lac repressor protein: This protein is involved in the regulation of lactose metabolism in bacteria.
- Cro protein: This protein is a transcriptional repressor that regulates gene expression in bacteriophages.
- CAP protein: This protein is involved in the regulation of glucose metabolism in bacteria.
- Trp repressor protein: This protein is a transcriptional repressor that regulates the biosynthesis of tryptophan in bacteria.
These proteins all contain a helix-turn-helix motif that is involved in binding to specific DNA sequences and regulating gene expression.
Helix-loop-helix
The helix-loop-helix (HLH) motif is a structural motif found in many proteins that are involved in DNA binding and gene regulation. The HLH motif consists of two alpha helices connected by a loop.

Here are some examples of proteins that contain a helix-loop-helix motif:
- Myc: This is a transcription factor that regulates the expression of genes involved in cell growth and division. Myc contains a basic HLH motif.
- Max: This is a protein that forms a dimer with Myc and regulates its activity. Max also contains a basic HLH motif.
- NeuroD: This is a transcription factor that regulates the development of neurons. NeuroD contains a basic HLH motif.
- Id proteins: These are inhibitors of DNA binding proteins that regulate the activity of other transcription factors. Id proteins contain a HLH motif but lack basic residues required for DNA binding.
- Twist: This is a transcription factor that regulates cell differentiation during embryonic development. Twist contains a HLH motif that is important for its dimerization with other transcription factors.
These proteins all contain a helix-loop-helix motif that is involved in DNA binding and gene regulation, and their specific functions depend on the interactions with other proteins and DNA sequences.
Zinc finger
Proteins known as zinc finger (ZF) proteins employ zinc as a structural cofactor. All ZFs have the property that their main amino acid sequence contains repetitions of four cysteine and/or histidine residues.
Each zinc finger domain is 30 amino acids long and forms a ββα configuration, where individual amino acids in the α-helix interact with three successive nucleotide bases in the major groove of DNA.
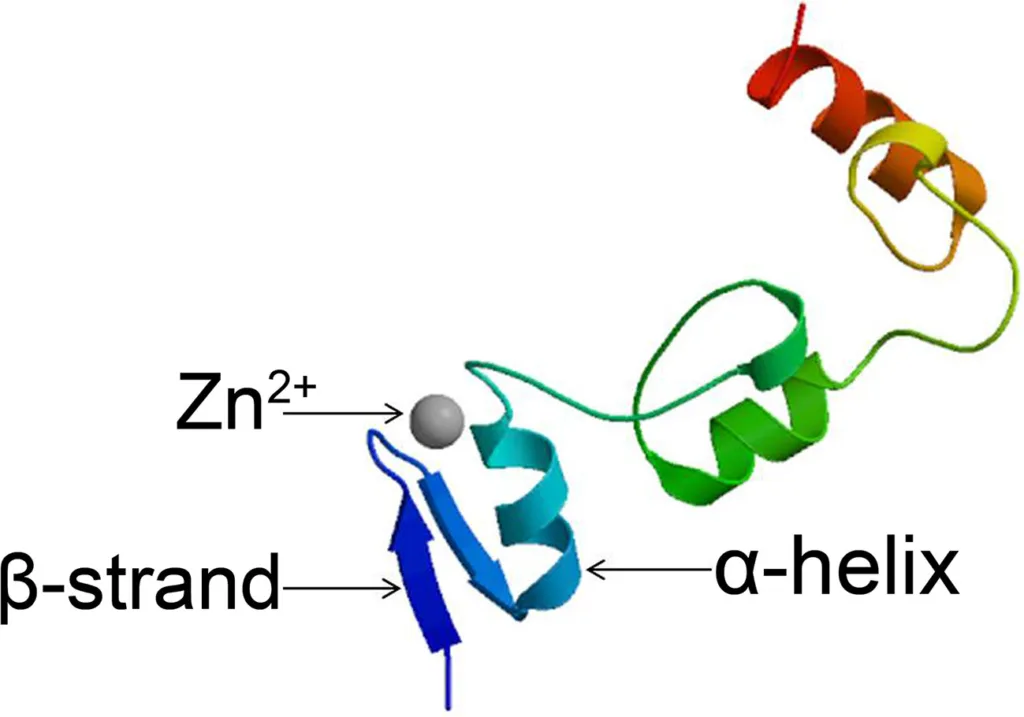
Here are some examples of proteins that contain zinc fingers:
- TFIIIA: This is a transcription factor that regulates the expression of ribosomal RNA genes. TFIIIA contains nine zinc fingers that bind to specific DNA sequences.
- Zinc finger nucleases: These are enzymes that can be engineered to cleave specific DNA sequences. Zinc finger nucleases contain a DNA cleavage domain and a DNA binding domain that consists of several zinc fingers.
- GATA transcription factors: These are transcription factors that are involved in the regulation of cell differentiation and development. GATA transcription factors contain two or more zinc fingers that bind to specific DNA sequences.
- Kruppel-like factors: These are transcription factors that regulate the expression of genes involved in cell proliferation, differentiation, and apoptosis. Kruppel-like factors contain three zinc fingers that bind to specific DNA sequences.
- Zinc finger antiviral protein (ZAP): This is a protein that inhibits the replication of many RNA viruses. ZAP contains four zinc fingers that are involved in RNA binding.
These proteins all contain zinc fingers that are involved in DNA or RNA binding and gene regulation, and their specific functions depend on the interactions with other proteins and DNA/RNA sequences.
Basic protein-leucine zipper
A structural motif known as the Basic protein-leucine zipper (bZIP) may be found in a variety of proteins that are involved in DNA binding and gene regulation. The bZIP motif is made up of a leucine zipper that enables dimerization of the protein and a basic region that binds to DNA.
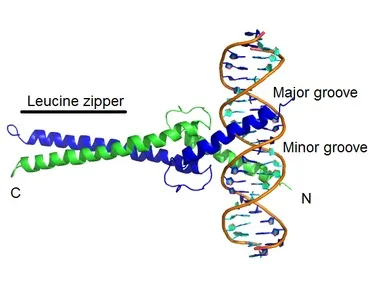
A few examples of proteins with the bZIP motif are as follows:
- c-Fos: It is a transcription factor that controls the proliferation and differentiation of cells. A crucial component of the dimerization of c-Fos and c-Jun is the presence of the bZIP motif.
- c-Jun: The control of cell development, differentiation, and death is regulated by this transcription factor. The dimerization of c-Jun and c-Fos depends on the presence of a bZIP motif in c-Jun.
- ATF-2: This transcription factor controls how cells respond to stress and how cell survival is regulated. An essential component of ATF-2’s ability to dimerize with other transcription factors is the presence of the bZIP motif.
- CREB: This transcription factor controls how genes are expressed in response to inputs like neurotransmitters and growth factors. A crucial component of CREB’s dimerization with other transcription factors is the presence of the bZIP motif.
- GCN4: This is a transcription factor that regulates the response to amino acid starvation in yeast. GCN4 contains a bZIP motif that is important for its dimerization with other transcription factors.
All of these proteins have the bZIP motif, which is involved in DNA binding and gene regulation, and each of these proteins has a unique function that is influenced by interactions with other proteins and DNA sequences.
β-sheet motifs
Many proteins have -sheet motifs, which are structural motifs made up of β-strands joined by loops or twists. These motifs are crucial for the stability, function, and folding of proteins.
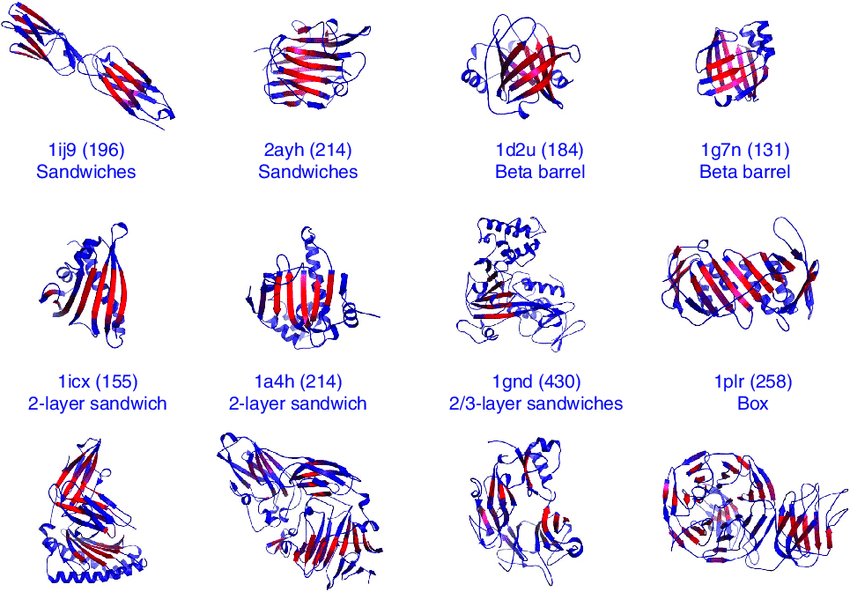
Here are some examples of β-sheet motifs:
- β-hairpin: Two antiparallel strands joined by a loop make up this -sheet pattern. Numerous proteins, including membrane proteins, enzymes, and antibodies, have -hairpins.
- β-meander: A number of strands joined by loops or twists make up this multiple β-stranded sheet pattern. Proteins with repeating amino acid sequences, like collagen and fibronectin, frequently include β-meanders.
- β-barrel: This is a β -sheet motif made up of several β -strands that come together to form a cylindrical or barrel-shaped structure. Many membrane proteins, including porins and enzymes, have β -barrels.
- β-propeller: This is a multi-stranded β-sheet motif that is structured in a propeller β-like or circular pattern. Many proteins involved in signaling and transport, such as lectins and proteases, have β-propellers.
- β-sandwich: The two β-sheets in this β-sheet motif are sandwiched between a hydrophobic core in a stacking arrangement. Many binding and recognition related proteins, such as antibodies and enzymes, include β-sandwiches.
These β-sheet motifs play crucial roles in protein structure and function, and their specific arrangements can influence protein stability, ligand binding, and catalytic activity.
References:
- https://www.frontiersin.org/articles/10.3389/fpls.2020.00115/full
- https://www.spandidos-publications.com/10.3892/wasj.2020.32
- https://www.sciencedirect.com/topics/neuroscience/transcription-factors