The translated proteins from mRNA undergo chemical or enzymatic modification before functioning in the cell which alters the amino acid sequence after its synthesis. This modification is known as post translational modification.
- Ribosomes synthesizes proteins and translate mRNA into polypetide chains which undergo post translational modification and forms the mature protein.
- The main function of post translational modification of protein is that it generates heterogeneity in proteins and helps in utilizing and functioning of each identical protein in different cell of the body.
- Post translational modification of protein increases the proteome functional diversity by the addition of functional groups, proteolytic cleavage of regulatory subunits or degradation of entire proteins.
- These modifications also influence proteins structure, stability, activity and substrate specificity.
- Post translational modification mostly occurs in the cells endoplasmic reticulum or sometimes in golgi bodies.
- This modification occurring at the peptide terminus of the amino acid helps in translocating proteins across biological membrane in prokaryotes and eukaryotes.
- They are also incorporated in various cellular and organelle membranes such as lysosomes, chloroplast, mitochondria and plasma membranes.
- Post translational modification also takes part in modifying the end product of expression that is important in biological processes and diseased conditions like heart disease, cancer, neurodegenerative diseases and diabetes.
Most proteins are modified shortly after completion of translation process to mediate proper protein folding or stability while other modifications occur after folding and catalytic activity that are activated or inactivated after localization is completed. Proteins go through post translational modification by cleavage and addition of functional groups through a mechanism of protein maturation or activation.
After completion of synthesis, proteins can be modified by various ways:
- Acetylation
- Glycosylation
- Lipidation
- Phosphorylation
- Methylation
- Proteolysis
- Ubiquitination
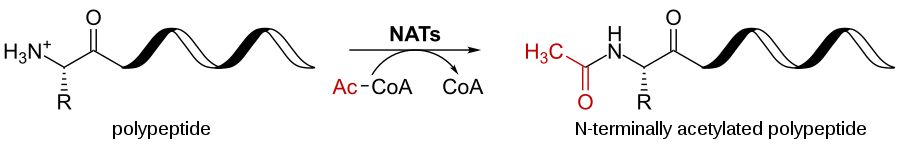
Acetylation
- During acetylation, the first amino group methionine in the polypeptide chain is replaced by acetyl group.
- The main function of acetylation is in gene expression that opens up the DNA for transcription.
- The histone protein is acetylated that condense DNA into chromatids and reduces the ability to fold and opens up.
- Acetylation occurs 80 to 90% of eukaryotic proteins.
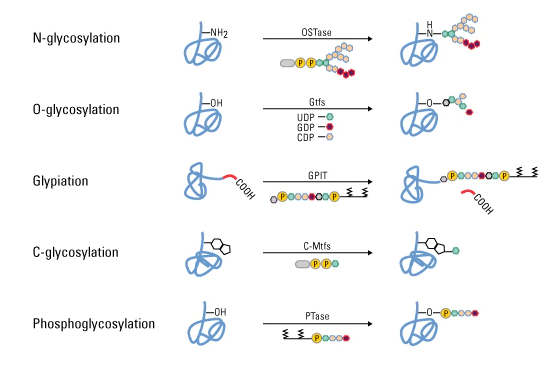
Glycosylation
- Protein glycosylation has a significant effect on protein folding, conformation, distribution, stability and activity.
- During glycosylation, carbohydrate is added to protein that forms from simple monosaccharide to highly complex branched polysaccharide.
- This change occur in cell surface receptor that helps in identifying different types of cell like in identification of ABO blood group.
- Four different red blood cells have four different proteins with different carbohydrate groups attached to them which help in determining the type of cells.
- Type of glycosylation is based on the nature of the sugar peptide bond and the oligosaccharide attached.
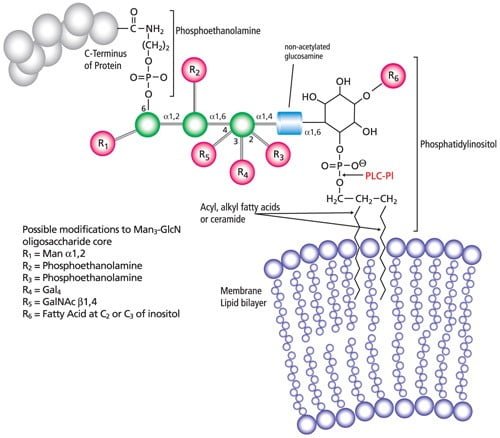
Lipidation – Post translational modifications
- Lipid molecule attaches to a protein on the cell membrane that usually occurs in endoplasmic reticulum or in golgi apparatus.
- C-terminal glycosyl phosphatidylinositol (GPI) anchor, N-terminal myristoylation, S-myristoylation and S-prenylation are four types of lipidation.
- One of the examples is GPI anchor that is a hydrophobic lipid which helps to attach proteins to the inner part of the cell membrane.
- This helps in the detection of GPI anchored proteins from membranes by gel separation and analysis by mass spectrometry.
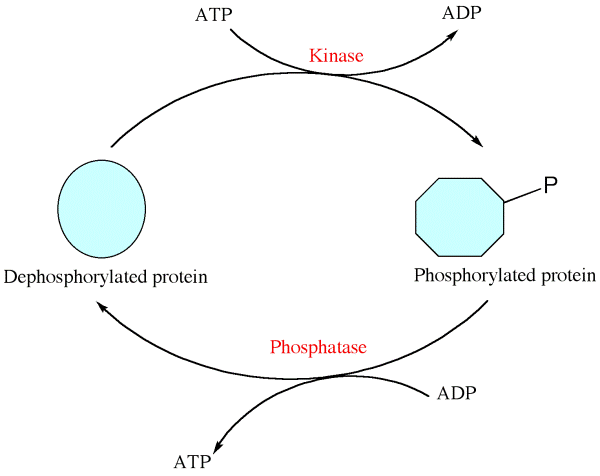
Phosphorylation – Post translational modifications
- Phosphorylation is the addition of phosphate group to certain amino acids like serine, threonine or tyrosine found on the polypeptide chain.
- Protein kinase enzyme is also involved in the catalysis of phosphorylation.
- Protein phosphorylation is a reversible process i.e, dephosphorylation occurs when phosphate group is removed from the protein or enzyme.
- Phosphorylation helps in the regulation of many cellular processes like cell cycle, cell growth, and apoptosis and in signal transduction pathways.
- Sodium – potassium pump is the best example of phosphorylation.
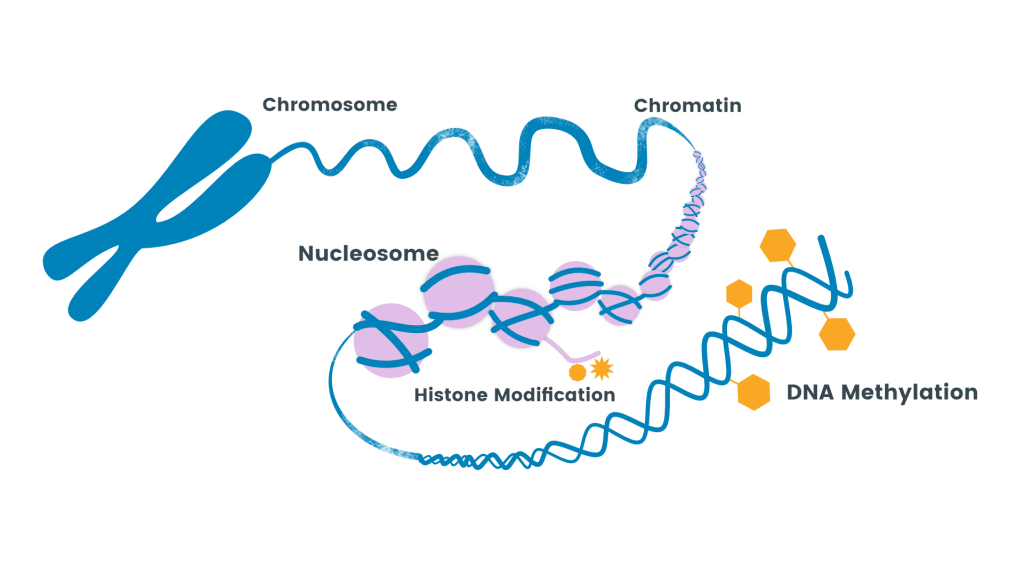
Methylation
- Methylation is the addition of methyl group to the amino acid by an enzyme methyltransferase.
- Methylation increases the hydrophobicity of the protein and can neutralize a negative amino acid charge by binding with carboxylic acids.
- Methylation and demethylation of histone protein help to turn on and off of certain genes as methylation is utilized in epigenetic regulation which helps in gene expression.
(Source: https://www.labclinics.com/en/role-dna-methylation-disease/)
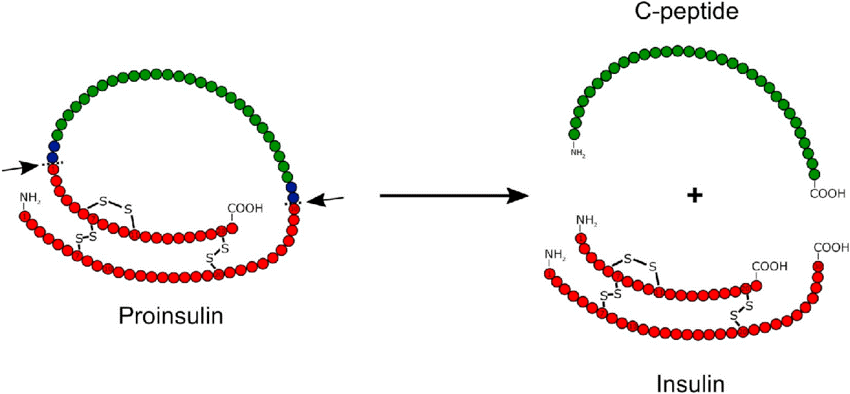
Proteolysis – Post translational modifications
- In proteolysis, protease enzyme cut certain peptide bonds of protein which gets synthesized in our body.
- Many digestive enzymes in the small intestine use proteolysis type of post translational modification.
- One of the most common examples of proteolysis is the production of insulin in our body which has to be cut twice before it gets activated.
Ubiquitination– Post translational modifications
- Ubiquitination is a process through which an ubiquitin molecule (an 8 kDa polytpeptide molecule) binds to a substrate protein and changes its way of functioning.
- Ubiquitination is catalysed by enzyme called as ubiquitin ligases that affects cellular process by regulating the degradation of proteins, activating and inactivating proteins and modulating protein-protein interactions.
Learn more
References:
- https://www.thermofisher.com/np/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/overview-post-translational-modification.html#:~:text=Overview%20of%20Post%2DTranslational%20Modifications%20(PTMs)&text=Protein%20post%2Dtranslational%20modifications%20(PTMs,or%20degradation%20of%20entire%20proteins.
- https://en.wikipedia.org/wiki/Post-translational_modification
- http://www.premierbiosoft.com/glycan/glossary/post-translational-modifications.html