Author: Binod G C
Introduction
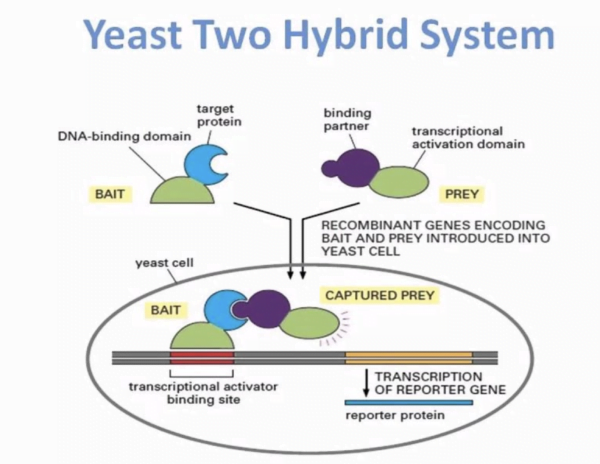
The Yeast Two-Hybrid (Y2H) Assay is a genetically engineered system designed to simplify the detection and evaluation of protein-protein interactions. In this system, a host organism, commonly yeast or bacteria, is modified to include three essential components. These components consist of a first protein fused to a DNA-binding domain with a known specificity, a second protein fused to a transcriptional activation domain that can interact with the first protein, forming a functional composite transcription factor. The activation of one or more reporter genes is dependent on the binding of this composite transcription factor. Over time, various adaptations of the two-hybrid system have been developed, and it has become a fundamental tool in proteomic research.
Principle of the Y2H Assay
The Y2H assay is based on the principle of reconstituting a transcription factor through the interaction of two proteins of interest. The bait protein is fused to a DNA binding domain (DBD), while the prey protein is fused to an activation domain (AD). The interaction between the bait and prey proteins leads to the reconstitution of a functional transcription factor, which activates the reporter gene expression.
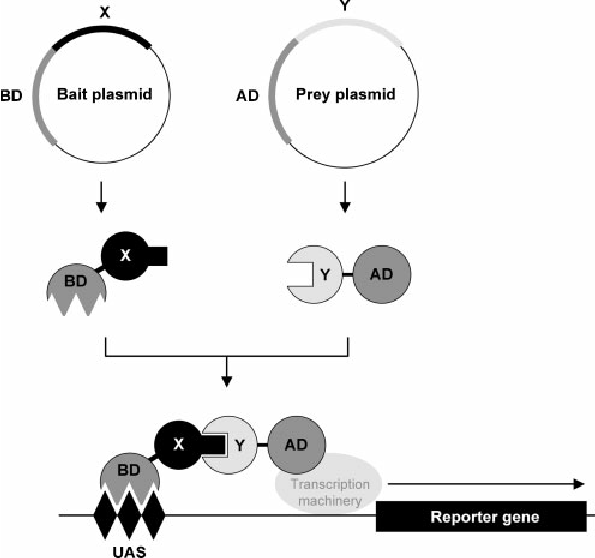
Procedure of Yeast Two-Hybrid (Y2H) Assay
The Y2H assay or Yeast Two-Hybrid (Y2H) Assay involves several steps:
- Design and construct the bait and prey plasmids:
Select the proteins of interest and clone their DNA sequences into separate plasmids. The bait plasmid contains the DBD fused to the bait protein, and the prey plasmid contains the AD fused to the prey protein.
- Transform yeast cells: Transform the bait and prey plasmids separately into compatible yeast strains. The bait plasmid is introduced into yeast cells with a reporter gene controlled by the DBD, while the prey plasmid is introduced into yeast cells with a reporter gene controlled by the AD.
- Mate bait and prey yeast strains: Mix the yeast cells carrying the bait and prey plasmids on selective agar plates to allow the mating process. Mating facilitates the interaction between the bait and prey proteins.
- Selective growth media: Transfer the mated yeast cells to selective growth media that only support the growth of cells with successful bait-prey interactions. This media lacks certain nutrients required by yeast cells, but the successful interaction reconstitutes the transcription factor and activates the reporter gene, allowing the cells to grow.
- Detection of reporter gene activation: Use appropriate methods to detect the activation of the reporter gene, depending on the reporter system employed. For example, assay colonies for enzyme activity if the reporter gene encodes a protein like β-galactosidase or use fluorescence microscopy/flow cytometry if the reporter gene encodes a fluorescent protein like GFP.
- Validation of interactions: Validate the identified interactions using additional techniques such as co-immunoprecipitation or other protein-protein interaction assays to confirm their specificity and reliability.
Variations of Y2H Assay
The Y2H assay can be adapted to study specific types of protein interactions:
1. Yeast 1-hybrid assay:
Examines protein-DNA interactions by replacing the prey protein with a target DNA sequence upstream of the reporter gene.
The yeast one-hybrid (Y1H) assay is a laboratory technique used to study the interactions between DNA and proteins within cells. It is an in vitro method that is based on the general principles of the yeast two-hybrid (Y2H) assay. The key distinction between the Y2H and Y1H assays is that the Y2H assay focuses on protein-protein interactions, whereas the Y1H assay specifically examines interactions between DNA and proteins. In the Y1H assay, the target proteins of interest are introduced into yeast cells, and the subsequent interactions between these proteins and DNA baits are assessed by monitoring the activation of reporter genes downstream.
2. Yeast 3-hybrid assay:
Studies protein interactions mediated by a third component, such as an RNA molecule or another protein, where the bait and prey proteins do not directly interact.
The yeast three-hybrid system is a modified version of the yeast two-hybrid (Y2H) technique. It serves as a valuable tool for studying specific RNA-protein interactions. This system involves three hybrid components that work together to facilitate the analysis of these interactions.
The first component consists of an RNA binding protein (RBD) fused to a DNA binding domain (DBD). The second component is another fusion protein, which combines a different RNA binding protein with a transcriptional activation domain (AD). The third component is an RNA molecule that acts as a bridge between the two fusion proteins. This RNA molecule provides specific RNA targets for the RNA binding proteins.
When these three components come together at a promoter site, the reporter gene is activated, albeit transiently. This activation leads to the production of reporter gene products, which can be detected and analyzed using simple biochemical or phenotypic assays. The yeast three-hybrid system thus enables the investigation of RNA-protein interactions of interest in a controlled experimental setup.
3. Split ubiquitin yeast 2-hybrid assay:
Used for identifying non-soluble membrane protein interactions. The bait and prey proteins are fused with different parts of the ubiquitin molecule, which reconstitutes upon interaction and activates reporter gene expression.
The ubiquitin protein is divided into two parts: the N-terminal domain (Nub) comprising amino acids 1-34, and the C-terminal domain (Cub) comprising amino acids 35-76. These halves are fused individually to proteins of interest, one to the bait protein and the other to the prey protein, resulting in hybrid-protein products. Additionally, the Cub peptide is linked to a reporter transcription factor.
When the Cub and Nub halves are brought into close proximity, they interact and reassemble to form a complete ubiquitin molecule. As a result, the reporter transcription factor is liberated from the Cub peptide due to the action of ubiquitin-specific proteases. The released transcription factor subsequently translocates into the nucleus and initiates the transcription of reporter genes, thereby facilitating the detection and measurement of protein-protein interactions.
Advantages and Limitations of Yeast Two-Hybrid (Y2H) Assay
The Y2H assay offers several advantages for studying protein-protein interactions:
- Straightforward methodology and fast results
- High-throughput screening capabilities
- Scalability to screen the entire proteome
- Adaptability to other model organisms for studying organism-specific interactions
However, it is important to consider the limitations of the Y2H assay:
- False positive and negative interactions that require validation
- Nucleus-dependent interaction, limiting the detection of interactions involving proteins in other cellular compartments
- Potential effects of overexpression and fusion proteins on protein function
- Need to confirm the expression and functionality of query proteins
Detailed protocol for Yeast Two-Hybrid (Y2H) Assay
Please note that all procedures described below should follow aseptic techniques, and yeast cultures should be grown at 30°C while Escherichia coli cultures should be grown at 37°C.
1. Construction and Transformation of the Bait Protein:
1.1 Charging and Equilibration of the ZipTipMC:
- Before conducting the interactor hunt, create plasmids that express the protein of interest fused with the bacterial protein LexA (or any other DNA-binding domain).
- These plasmids will be introduced into a yeast reporter strain to assess the suitability of the bait proteins for library screening.
- Insert the DNA encoding the protein of interest into the polylinker of pMW103 to generate a fusion with LexA. Prepare competent yeast cells from a colony of SKY48.
Transform the competent yeast cells with different combinations of plasmids (100-500 ng each):
a) pBait + pMW109 (test for activation)
b) pEG202-hsRPB7 + pMW109 (weak positive control for activation)
c) pSH17-4 + pMW109 (strong positive control for activation)
d) pEG202-Ras + pMW109 (negative control for activation)
- Plate each transformed mixture on Glu/CM -Ura-His dropout plates and incubate for 2 to 3 days to select yeast colonies containing the transformed plasmids.
2. Assessment of Bait Activation and Expression:
2.1 Replica Technique/Gridding Yeast: Assessing Activation of Auxotrophic Reporter:
- Evaluate at least six independent colonies for the activation phenotype of auxotrophic and colorimetric reporters for each combination of plasmids.
- Transfer yeast cells from individual patches on the master plate to various selective media using a sterile toothpick or a high-throughput transfer technique.
- Select six yeast colonies (1 to 2 mm diameter) from each transformation plate (a-d) and position the tips supported by an insert grid placed over a 96-well microtiter plate.
- Gently swirl the plate to suspend the yeast cells, remove the tape holding the grid, and lift the insert grid, removing all the tips at once.
Use a replicator to plate yeast suspensions on the following plates:
a) Glu/CM -Ura-His (new master plate)
b) Gal-Raff/CM -Ura-His-Leu
c) Gal-Raff/CM -Ura-His
- Incubate the plates for up to 4 days and store the Glu/CM -Ura-His master plate at 4°C. Check the growth of yeast colonies on the selective plates.
- Positive controls (b and c) should exhibit growth within 1-2 days and 4 days, respectively.
- The negative control (d) should not show any growth. If the tested bait (a) does not grow within this timeframe, it is likely suitable for library screening.
2.2 Assessment of Colorimetric Reporter Activation:
- After 24 to 30 hours of plating, cover the Gal-Raff/CM -Ura-His plate with X-Gal agarose.
- Gently overlay each plate with chloroform and fully cover the colonies for 5 minutes.
- Then, briefly overlay the plates with approximately 5 mL of chloroform and let them dry uncovered for an additional 5 minutes at 37°C or 10 minutes in a chemical hood.
- Cover the plates with approximately 10 mL of X-Gal agarose, ensuring that all yeast spots are completely covered.
- Incubate the plates at 30°C and observe for color development. Blue colonies indicate the expression and activation of the bait protein fusion.
3. Construction and Transformation of a Prey Library:
3.1 Preparation and Transformation of the Prey Library:
- Obtain a cDNA library cloned into a prey vector.
- Amplify the cDNA library plasmids in E. coli, purify the DNA, and determine the concentration.
- Linearize the prey library plasmids by digesting them with a restriction enzyme.
- Prepare competent yeast cells (e.g., Y187).
- Transform the linearized prey library plasmids into the competent yeast cells using the lithium acetate method.
- Plate the transformed yeast cells on Glu/CM -Ura dropout plates and incubate for 2 to 3 days to select yeast colonies containing the prey library plasmids.
4. Mate Transformation:
4.1 Preparation and Mating of Bait and Prey Yeast Strains:
- Prepare saturated cultures of the bait strain (e.g., Y2HGold) and the prey library strain (e.g., Y187) in liquid Glu/CM -Ura medium.
- Dilute the saturated cultures to OD600 = 0.5 in 50 mL of YPDA medium and incubate at 30°C with shaking at 200 rpm.
- Mix 2 mL of the bait culture with 2 mL of the prey library culture in a 15 mL conical tube.
- Centrifuge the cells at 3,000 x g for 5 minutes, discard the supernatant, and resuspend the cell pellet in 200 μL of sterile water.
- Spread the suspension evenly on a YPDA plate and incubate at 30°C for 24 hours.
5. Selective Screening and Confirmation of Interactions:
5.1 Selective Screening for Interactions:
- After 24 hours of mating, replica plate the mated cells onto selective medium plates using a sterile 96-pin replicator.
- Incubate the plates at 30°C for 2 to 4 days and observe the growth of yeast colonies.
- Assess potential positive interactions based on the growth of yeast colonies on selective medium plates.
5.2 Confirmation of Interactions:
- For positive interactions, pick yeast colonies from the selective medium plates and streak them onto Glu/CM -Ura-His plates.
- Incubate the plates at 30°C for 2 to 3 days and check for the growth of yeast colonies.
- Perform appropriate control experiments to validate the specificity of the interactions.
6. Other Controls and Considerations:
- Include suitable positive and negative controls throughout the assay to ensure the reliability of the results.
- Employ stringent washes and appropriate stringency controls during the assay to minimize false positives.
- Validate positive interactions using additional techniques such as co-immunoprecipitation, Western blotting, or fluorescence microscopy.
This protocol serves as a general guideline for the Yeast Two-Hybrid (Y2H) Assay. However, specific details and conditions may vary depending on the system and reagents used. It is recommended to refer to the literature and manufacturer’s instructions for the specific reagents and vectors employed.
Conclusion
The Yeast Two-Hybrid (Y2H) Assay is a powerful technique for detecting and studying protein-protein interactions. It provides researchers with a valuable tool to investigate the complex network of interactions that occur within cells. While it has limitations, its simplicity and high-throughput capabilities make it an essential component of many molecular biology studies. Researchers should be aware of the assay’s advantages and limitations and take appropriate steps to ensure the accuracy of the obtained results.
References
- Fields, S., & Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature, 340(6230), 245-246.
- Vidalain, P. O., & Boxem, M. (2006). GeTools: analysis of interactomics and association networks with multiple tools. Bioinformatics, 22(3), 376-377.
- Stynen, B., & Tournu, H. (2018). The role of ERMES in mitochondrial biology: expanding the crista junctions. Biochemical Society Transactions, 46(4), 827-839.
- Klug, L., et al. (2019). Analysis of protein-protein interactions using the yeast two-hybrid assay. Methods in Molecular Biology, 2004, 3-20.
- Eyckerman, S., & Lemmens, I. (2006). Studying protein-protein interactions: progress, pitfalls, and solutions. Journal of Proteome Research, 5(4), 935-944.
- Gavin, A. C., et al. (2006). Proteome survey reveals modularity of the yeast cell machinery. Nature, 440(7084), 631-636.
- Miller, J. P., et al. (2015). Yeast two-hybrid screens imply involvement of Fanconi anemia proteins in transcription regulation, cell signaling, oxidative metabolism, and cellular transport. Experimental Cell Research, 337(2), 274-286.
Learn more