History
Edwin Southern, the inventor of Southern blotting started a trend to his invention after him. It was introduced as a technique to detect particular sequence of DNA in DNA samples. He first published the article in 1975.
Southern integrated three innovations to create the Southern blot – restriction endonucleases, gel electrophoresis and blotting through methods.DNA fragments were differentiated using electrophoresis based on size, then transferred to a membrane and hybridized with a radio labeled DNA probe.
Definition
Southern blotting is a molecular biology technique used for DNA detection, characterization, and quantification.
An example of RFLP(restriction fragment length polymorphism), southern blotting can be defined as an analytical technique for identifying specific sequences of DNA by separating fragments on a gel and transferring them to a second medium (carrier membrane) on which hybridization testing may be carried out. During southern blotting, the DNA fragments are immobilized as a result, the membrane carries a semi-permanent reproduction of the banding pattern of the gel. The DNA are then exposed to hybridization analysis allowing bands with sequence resemblance to a labeled probe to be identified.
There are different types of membrane, transfer buffer and transfer methods to set up a southern blot. The most common and popular membranes are made of nitrocellulose, uncharged nylon positively charged nylon but they are interchangeable depending on the applications.
Principle of Western Blotting
It is based on the principle of transfer of separated DNA fragments to a carrier membrane (usually nitrocellulose) using gel electrophoresis and subsequent identification of specific DNA fragmentsby labelled probe hybridization. Hybridization is a technique in which a double stranded DNA molecule is formed in between a single stranded DNA probe and a target single stranded DNA. The probes are labeled with a marker and complementary to the target DNA as a result we can detect one molecule of target in a mixture of millions after hybridization as the reactions are specific.
Procedure of Southern Blotting
- Extract and purify DNA from cells
- We separate the DNA to be tested from the rest of the cellular material in the nucleus.
- We then incubate the specimen with detergent to promote cell lysis (frees cellular proteins and DNA).
- Proteins are removed through organic and non-organic extraction.
- We then use alcohol precipitation to purify the DNA from the solution.
- Visible DNA fibers are removed and suspended in buffer.
2. DNA fragmentation
- We use restriction endonuclease enzyme to break long nucleotide sequence into smaller fragments for purification or identification process.
- Each restriction enzymes are validated with universal buffers (L, M, H, K, or T (+BSA)) and supplied with recommended buffer.
- Before appropriate DNA concentration and establishing a restriction digestion with preferable enzymes, we keep reagent necessary for digestion process on ice.
- The components are then added into a PCR tube and mixedby absorbing the contents with the help of pipette slowly avoiding formation of any bubbles.
- PCR amplifies the number of fragments of DNA obtained from the restriction digest which are easily separated using gel electrophoresis.
3. Gel electrophoresis
- Sorts the complex mixture of DNA fragments according to size.
- The percentage and size of the gel to be used must be determined.
- Gels consist of microscopic pores and are solid (usually agarose or polyacrimide).
Generally, 0.7 – 2% gel is considered to be adequate for most of the applications. - Nucleic acids have negative charge and move from left to right. The large molecules are held up while smaller ones move faster causing separation by size.
- Gels are stained with ethidium bromide to permit photography under UV light.
- A single band form is given to intact high quality DNA (small amount of degradation is tolerable).
4. Denature DNA
- DNA obtained are double stranded in nature.
- Alkalis are used to denature the restriction fragments in the gel that makes double stranded DNA to become single stranded.
- To avoid re-hybridization, we use NaCl so that DNA is neutralized.
5. Blotting
- Transfer DNA from the gel to solid support (carrier membrane).
- We dry the blot (around 80°C) or use UV radiation to make it permanent.
6. Hybridization
- The membrane bounded with DNA are incubated after adding the labelled probe.
- Usually requires 1-16 hours depending on the complexity of the probe and concentration.
- The probe then binds with complementary DNA on the membrane with the help of BSA or casein (blocks all other non-specific binding).
7. Washing
- Despite using blocking agents, some excess probe binds to the membrane.
- Wash buffers containing NaCl and detergent washes away the excess probe.
8. Autoradiography
- The particles are exposed to X-ray film when we use radioactive probe or fluorescent probe.
- If a chromogenic detection method is used, we can see development of color on the membrane.
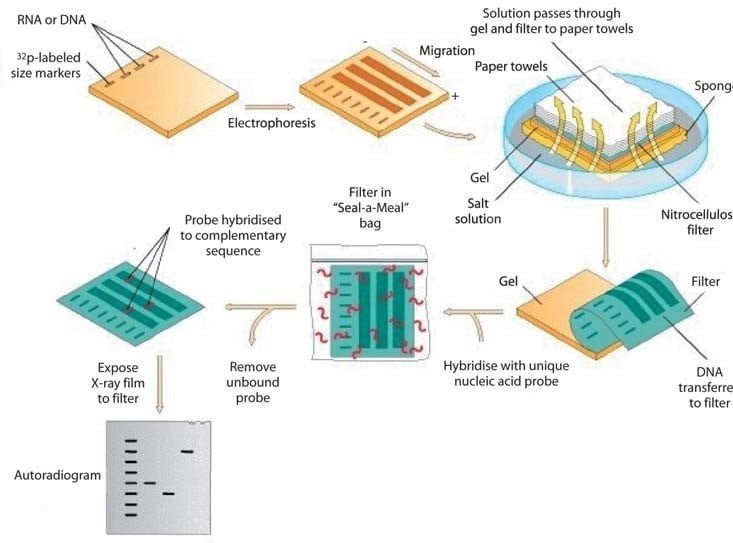
Figure adapted from: The mycology of the Basidiomycetes
Applications of Southern Blotting
- Identify specific DNA from a DNA sample.
- Identification of viral and bacterial infections.
- Important in the study of gene mutation, deletion and rearrangements.
- DNA fingerprinting (maternity and paternity analysis, forensic studies and personal identification).
- Diagnosis of neonatal and genetic diseases including cancer.
- Discovery of RFLP (restriction fragment length polymorphism) to map crucial genomes.
Advantage and Disadvantages
Advantages
- Less degraded compared with protein and mRNA as DNA are very stable.
- Effective way to detect specific DNA sequence from large complex samples.
- Increased sensitivity of fragments detection because of probe label used for amplifying signals.
- Only way to diagnose FSHD (Facioscapulohumerals Muscular Dystrophy).
Disadvantages
- Not applicable in routine diagnostic setting.
- Time consuming and requires large amount of DNA.
- Does not allow morphologic preservation of tissue so historic evaluation features are not available.
References
- Terry Brown (1993), Analysis of DNA Sequences by Blotting andHybridization
- Terence A Brown (2001), Southern Blotting and Related DNA Detection Techniques
- DaidreeTofano, Ilse R. Wiechers, Robert Cook-Deegan (2006), Edwin Southern, DNA blotting, and microarray technology: A case study of the shifting role of patents in academic molecular biology
- Ed Southern (2006), Southern blotting protocols
- Ian A. Hood (2006),The mycology of the Basidiomycetes
- Daniela Furrer, Francois Sanschargin, Simon Jacob, Caroline Diorio (2015), Advantages and Disadvantages of Technologies for HER2 Testing in Breast Cancer Specimens
- Manu Tomar(2016), Types of Blotting, Research & Reviews: Journal of Pharmaceutics and Nanotechnology